library(tidymodels)
library(forested)
# Set a seed
set.seed(123)
# IntitalizeInitialize Split
forested_split <- initial_split(forested, prop = 0.8)
forested_train <- training(forested_split)
forested_test <- testing(forested_split)
# Build Resamples
forested_folds <- vfold_cv(forested_train, v = 10)
# Set a model specification with mode (default engine)
dt_mod <- decision_tree(cost_complexity = 0.0001, mode = "classification")
# Bare bones workflow & fit
forested_wflow <- workflow(forested ~ ., dt_mod)
forested_fit <- fit(forested_wflow, forested_train)
# Extracting Predictions:
augment(forested_fit, new_data = forested_train)
#> # A tibble: 5,685 × 22
#> .pred_class .pred_Yes .pred_No forested year elevation eastness northness
#> <fct> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl>
#> 1 No 0.0114 0.989 No 2016 464 -5 -99
#> 2 Yes 0.636 0.364 Yes 2016 166 92 37
#> 3 No 0.0114 0.989 No 2016 644 -85 -52
#> 4 Yes 0.977 0.0226 Yes 2014 1285 4 99
#> 5 Yes 0.977 0.0226 Yes 2013 822 87 48
#> 6 Yes 0.808 0.192 Yes 2017 3 6 -99
#> 7 Yes 0.977 0.0226 Yes 2014 2041 -95 28
#> 8 Yes 0.977 0.0226 Yes 2015 1009 -8 99
#> 9 No 0.0114 0.989 No 2017 436 -98 19
#> 10 No 0.0114 0.989 No 2018 775 63 76
#> # ℹ 5,675 more rows
#> # ℹ 14 more variables: roughness <dbl>, tree_no_tree <fct>, dew_temp <dbl>,
#> # precip_annual <dbl>, temp_annual_mean <dbl>, temp_annual_min <dbl>,
#> # temp_annual_max <dbl>, temp_january_min <dbl>, vapor_min <dbl>,
#> # vapor_max <dbl>, canopy_cover <dbl>, lon <dbl>, lat <dbl>, land_type <fct>Lecture 19
Evaluation & Tuning
Recaping Day 1 - 4
Evaluation
- So far we have used
metricsandcollect_metricsto evaluate and compare models - The default metrics for classification problems are:
accuracy,brier_score,roc_auc- All classification metrics stem from the classic confusion matrix
- The default metrics for regression problems are:
rsq,rmse,mae- The majority of these are measures of correlation and residual error
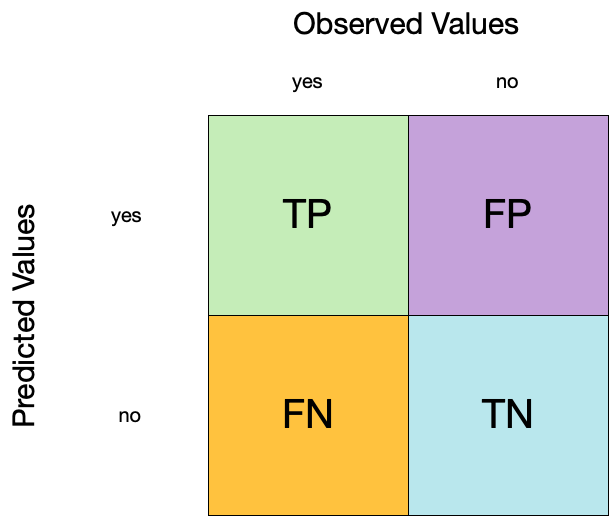
Classification:
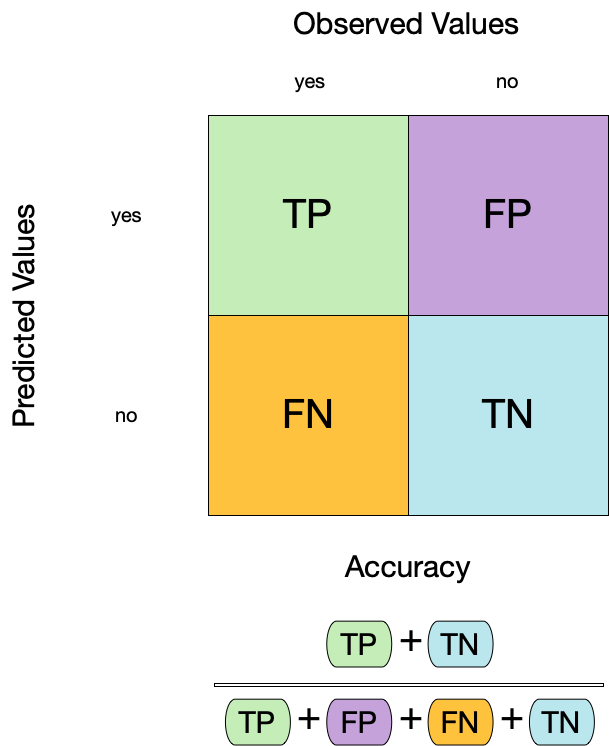
Confusion matrix ![]()
- A confusion matrix is a table that describes the performance of a classification model on a set of data for which the true values are known.
- It counts the number of accurate and false predictions, separated by the truth state
What to do with a confusion matrix
1. Accuracy ![]()
- Accuracy is the proportion of true results (both true positives and true negatives) among the total number of cases examined.
- It is a measure of the correctness of the model’s predictions.
- It is the most common metric used to evaluate classification models.
2. Sensitivity ![]()
- Sensitivity is the proportion of true positives to the sum of true positives and false negatives.
- It is useful for identifying the presence of a condition.
- It is also known as the true positive rate, recall, or probability of detection.
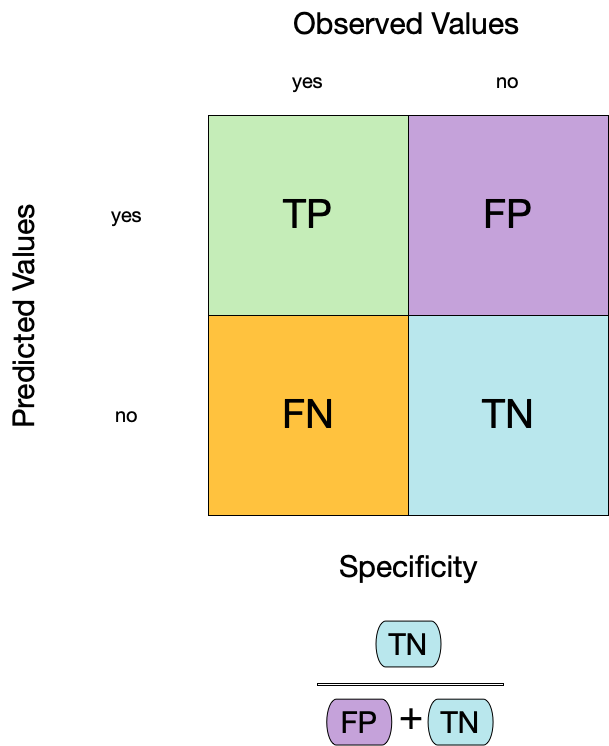
3. Specificity ![]()
- Specificity is the proportion of true negatives to the sum of true negatives and false positives.
- It is useful for identifying the absence of a condition.
- It is also known as the true negative rate, and is the complement of sensitivity.
Two class data
These metrics assume that we know the threshold for converting “soft” probability predictions into “hard” class predictions.
For example, if the predicted probability of a tree is 0.7, we might say that the model predicts “Tree” for that observation.
If the predicted probability of a tree is 0.4, we might say that the model predicts “No tree” for that observation.
The threshold is the value that separates the two classes.
The default threshold is 0.5, but this can be changed.
Is a 50% threshold good?
What happens if we say that we need to be 80% sure to declare an event?
- sensitivity ⬇️, specificity ⬆️
What happens for a 20% threshold?
- sensitivity ⬆️, specificity ⬇️
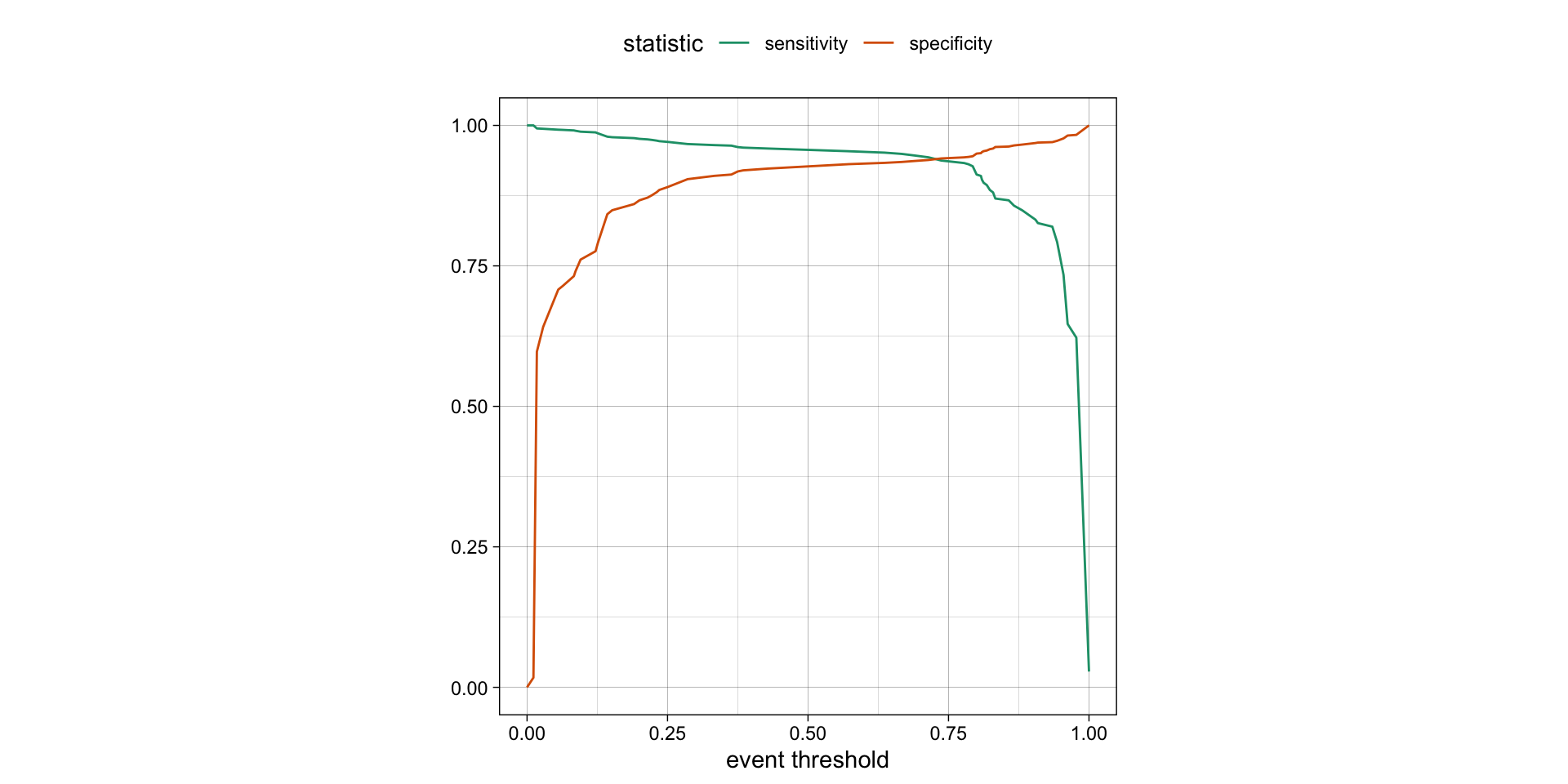
Varying the threshold
- The threshold can be varied to see how it affects the sensitivity and specificity of the model.
- This is done by plotting the sensitivity and specificity against the threshold.
- The threshold is varied from 0 to 1, and the sensitivity and specificity are calculated at each threshold.
- The plot shows the trade-off between sensitivity and specificity at different thresholds.
- The threshold can be chosen based on the desired balance between sensitivity and specificity.
ROC curves
For an ROC (receiver operator characteristic) curve, we plot
- the false positive rate (1 - specificity) on the x-axis
- the true positive rate (sensitivity) on the y-axis
with sensitivity and specificity calculated at all possible thresholds.

ROC curves
The ROC AUC is the probability that a randomly chosen positive instance is ranked higher than a randomly chosen negative instance.
The ROC AUC is a measure of how well the model separates the two classes.
The ROC AUC is a number between 0 and 1.
ROC AUC = 1 💯
- perfect classification
ROC AUC = 0.5 😐
- random guessing
ROC AUC < 0.5 😱
- worse than random guessing
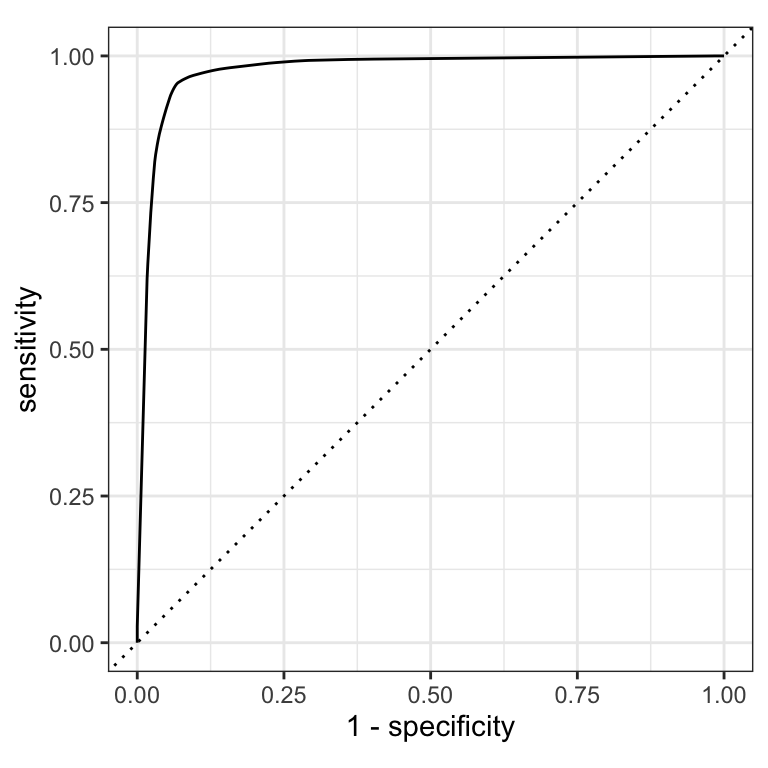
ROC curves ![]()
(A ROC AUC of 0.7-0.8 is considered acceptable, 0.8-0.9 is good, and >0.9 is excellent.)
# Assumes _first_ factor level is event; there are options to change that
augment(forested_fit, new_data = forested_train) |>
roc_curve(truth = forested, .pred_Yes) |>
dplyr::slice(1, 20, 50)
#> # A tibble: 3 × 3
#> .threshold specificity sensitivity
#> <dbl> <dbl> <dbl>
#> 1 -Inf 0 1
#> 2 0.235 0.885 0.972
#> 3 0.909 0.969 0.826
augment(forested_fit, new_data = forested_train) |>
roc_auc(truth = forested, .pred_Yes)
#> # A tibble: 1 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 roc_auc binary 0.975Brier score
The Brier score is a measure of how well the predicted probabilities of an event match the actual outcomes.
It is the mean squared difference between predicted probabilities and actual outcomes.
The Brier score is a number between 0 and 1.
The Brier score is analogous to the mean squared error in regression models:
Brier = 1 😱
- predicted probabilities are completely wrong
Brier = 0.5 😐
- bad
Brier < 0.25
- acceptable
Brier = 0 💯
- predicted probabilities are perfect
\[ Brier_{class} = \frac{1}{N}\sum_{i=1}^N\sum_{k=1}^C (y_{ik} - \hat{p}_{ik})^2 \]
Brier score
Separation vs calibration
The ROC captures separation. - The ROC curve shows the trade-off between sensitivity and specificity at different thresholds.
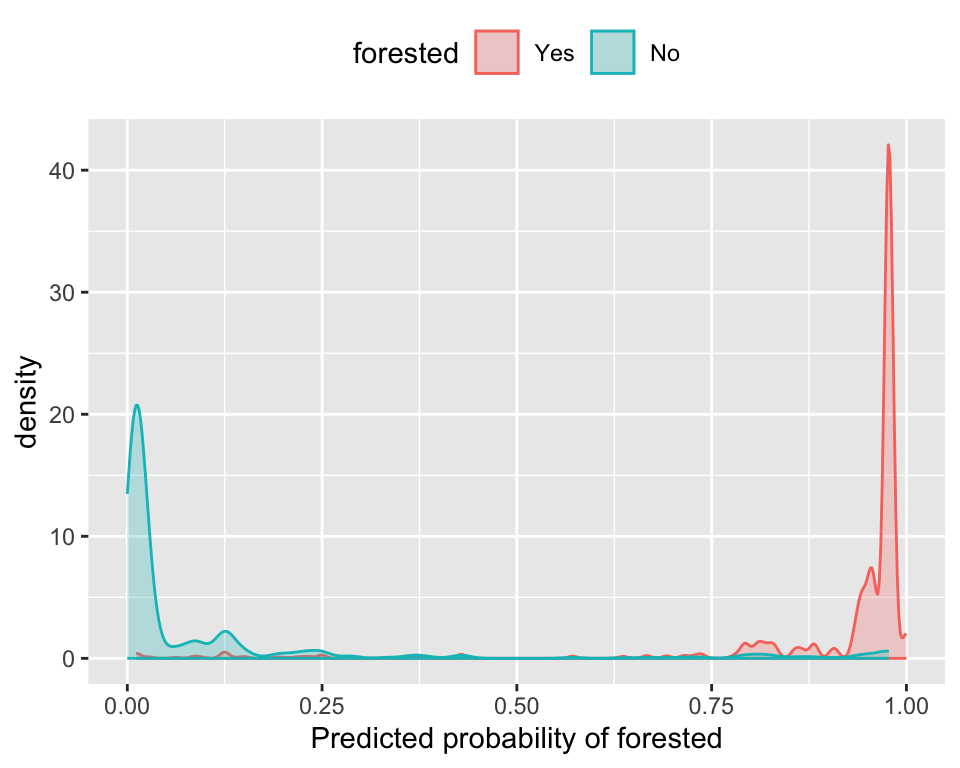
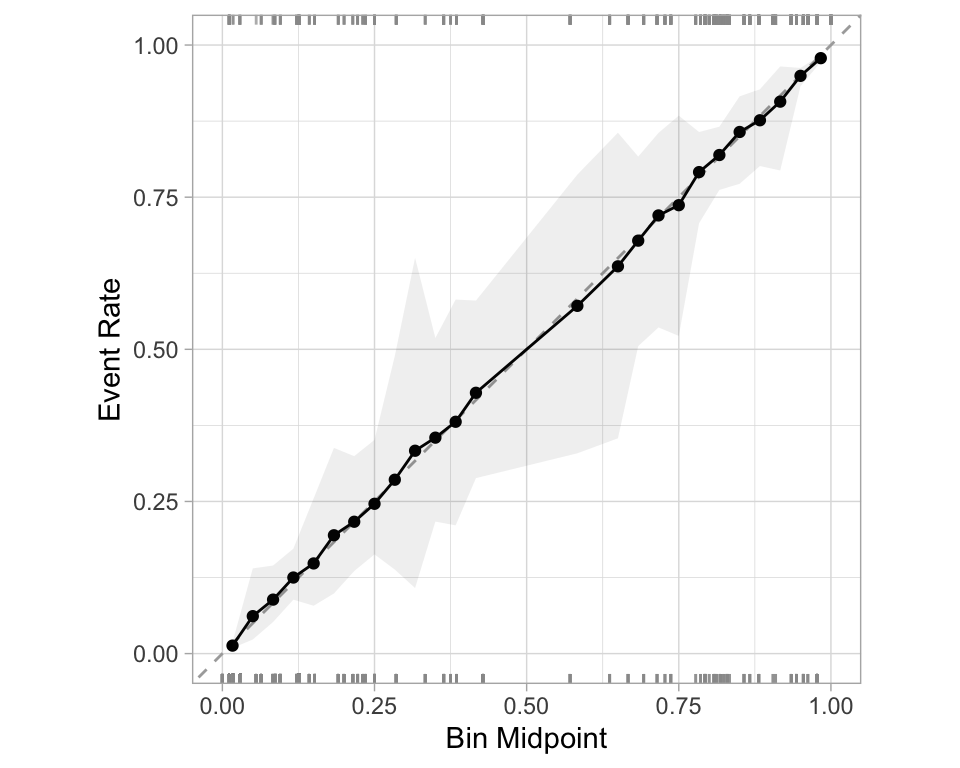
The Brier score captures calibration. - The Brier score is a measure of how well the predicted probabilities of an event match the actual outcomes.
- Good separation: the densities don’t overlap.
- Good calibration: the calibration line follows the diagonal.
Calibration plot: We bin observations according to predicted probability. In the bin for 20%-30% predicted prob, we should see an event rate of ~25% if the model is well-calibrated.
Regression
R-squared (\(R^2\))
- Measures how well the model explains the variance in the data.
- Formula:
\[
R^2 = 1 - \frac{SS_{res}}{SS_{tot}}
\] where:
- \(SS_{res}\) is the sum of squared residuals.
\(SS_{tot}\) is the total sum of squares.
Values range from \(-\infty\) to 1.
- 1: Perfect fit
- 0: No improvement over mean prediction
- Negative: Worse than just using the mean
- 1: Perfect fit
Root Mean Squared Error (RMSE)
- Measures the model’s prediction error in the same unit as the dependent variable.
- Formula:
\[ RMSE = \sqrt{\frac{1}{n} \sum_{i=1}^{n} (y_i - \hat{y}_i)^2} \]
where: - \(y_i\) is the actual value. - \(\hat{y}_i\) is the predicted value.
- Penalizes large errors more than MAE.
- Lower RMSE indicates better model performance.
Mean Absolute Error (MAE)
- Measures average absolute errors between predictions and actual values.
- Formula:
\[ MAE = \frac{1}{n} \sum_{i=1}^{n} |y_i - \hat{y}_i| \]
- Less sensitive to outliers than RMSE.
- Lower MAE indicates better model performance.
Comparing Metrics
| Metric | Interpretation | Sensitivity to Outliers |
|---|---|---|
| \(R^2\) | Variance explained | Moderate |
| RMSE | Penalizes large errors more | High |
| MAE | Average error size | Low |
Choosing the Right Metric
- Use \(R^2\) to assess model fit and explainability.
- Use RMSE if large errors are especially undesirable.
- Use MAE for a more interpretable, robust metric.
Metrics Selection model performance ![]()
We can use metric_set() to combine multiple calculations into one
forested_metrics <- metric_set(accuracy, specificity, sensitivity)
augment(forested_fit, new_data = forested_train) |>
forested_metrics(truth = forested, estimate = .pred_class)
#> # A tibble: 3 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 accuracy binary 0.944
#> 2 specificity binary 0.931
#> 3 sensitivity binary 0.954Metrics and metric sets work with grouped data frames!
augment(forested_fit, new_data = forested_train) |>
group_by(tree_no_tree) |>
accuracy(truth = forested, estimate = .pred_class)
#> # A tibble: 2 × 4
#> tree_no_tree .metric .estimator .estimate
#> <fct> <chr> <chr> <dbl>
#> 1 Tree accuracy binary 0.946
#> 2 No tree accuracy binary 0.941
augment(forested_fit, new_data = forested_train) |>
group_by(tree_no_tree) |>
specificity(truth = forested, estimate = .pred_class)
#> # A tibble: 2 × 4
#> tree_no_tree .metric .estimator .estimate
#> <fct> <chr> <chr> <dbl>
#> 1 Tree specificity binary 0.582
#> 2 No tree specificity binary 0.974Note
The specificity for "Tree" is a good bit lower than it is for "No tree".
So, when this index classifies the plot as having a tree, the model does not do well at correctly identifying the plot as non-forested when it is indeed non-forested.
Recap
Previously - Setup ![]()
library(tidyverse)
# Ingest Data
# URLs for COVID-19 case data and census population data
covid_url <- 'https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-states.csv'
pop_url <- '/Users/mikejohnson/Downloads/co-est2023-alldata.csv'
#pop_url <- 'https://www2.census.gov/programs-surveys/popest/datasets/2020-2023/counties/totals/co-est2023-alldata.csv'
# Clean Census Data
census = readr::read_csv(pop_url) |>
filter(COUNTY == "000") |> # Filter for state-level data only
mutate(fips = STATE) |> # Create a new FIPS column for merging
select(fips, contains("2021")) # Select relevant columns for 2021 data
# Process COVID-19 Data
state_data <- readr::read_csv(covid_url) |>
group_by(fips) |>
mutate(
new_cases = pmax(0, cases - dplyr::lag(cases)), # Compute new cases, ensuring no negative values
new_deaths = pmax(0, deaths - dplyr::lag(deaths)) # Compute new deaths, ensuring no negative values
) |>
ungroup() |>
left_join(census, by = "fips") |> # Merge with census data
mutate(
m = month(date), y = year(date),
season = case_when( # Define seasons based on month
m %in% 3:5 ~ "Spring",
m %in% 6:8 ~ "Summer",
m %in% 9:11 ~ "Fall",
m %in% c(12, 1, 2) ~ "Winter"
)
) |>
group_by(state, y, season) |>
mutate(
season_cases = sum(new_cases, na.rm = TRUE), # Aggregate seasonal cases
season_deaths = sum(new_deaths, na.rm = TRUE) # Aggregate seasonal deaths
) |>
distinct(state, y, season, .keep_all = TRUE) |> # Keep only distinct rows by state, year, season
ungroup() |>
select(state, contains('season'), y, POPESTIMATE2021, BIRTHS2021, DEATHS2021) |> # Select relevant columns
drop_na() |> # Remove rows with missing values
mutate(logC = log(season_cases +1)) # Log-transform case numbers for modelingPreviously - Data Usage ![]()
Previously - Feature engineering ![]()
Optimizing Models via Tuning Hyperparameter
Tuning parameters
Some model or preprocessing parameters cannot be estimated directly from the data.
Try different values and measure their performance.
- Find good values for these parameters.
- Once the value(s) of the parameter(s) are determined, a model can be finalized by fitting the model to the entire training set.
Tagging parameters for tuning ![]()
With tidymodels, you can mark the parameters that you want to optimize with a value of tune().
The function itself just returns… itself:
Boosted Trees
In last weeks live demo, a boosted tree proved most effective.
Boosted Trees are popular ensemble methods that build a sequence of tree models.
Each tree uses the results of the previous tree to better predict samples, especially those that have been poorly predicted.
Each tree in the ensemble is saved and new samples are predicted using a weighted average of the votes of each tree in the ensemble.
Boosted Tree Tuning Parameters
Some possible parameters:
mtry: The number of predictors randomly sampled at each split (in \([1, ncol(x)]\) or \((0, 1]\)).trees: The number of trees (\([1, \infty]\), but usually up to thousands)min_n: The number of samples needed to further split (\([1, n]\)).learn_rate: The rate that each tree adapts from previous iterations (\((0, \infty]\), usual maximum is 0.1).stop_iter: The number of iterations of boosting where no improvement was shown before stopping (\([1, trees]\))
Boosted Tree Tuning Parameters ![]()
![]()
b_mod <-
boost_tree(trees = tune(), learn_rate = tune()) |>
set_mode("regression") |>
set_engine("xgboost")
(b_wflow <- workflow(rec, b_mod))
#> ══ Workflow ════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: boost_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
#> 4 Recipe Steps
#>
#> • step_rm()
#> • step_dummy()
#> • step_scale()
#> • step_center()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
#> Boosted Tree Model Specification (regression)
#>
#> Main Arguments:
#> trees = tune()
#> learn_rate = tune()
#>
#> Computational engine: xgboostOptimize tuning parameters
The main two strategies for optimization are:
Grid search 💠 which tests a pre-defined set of candidate values
Iterative search 🌀 which suggests/estimates new values of candidate parameters to evaluate
1. Grid search
Most basic (but very effective) way to tune models
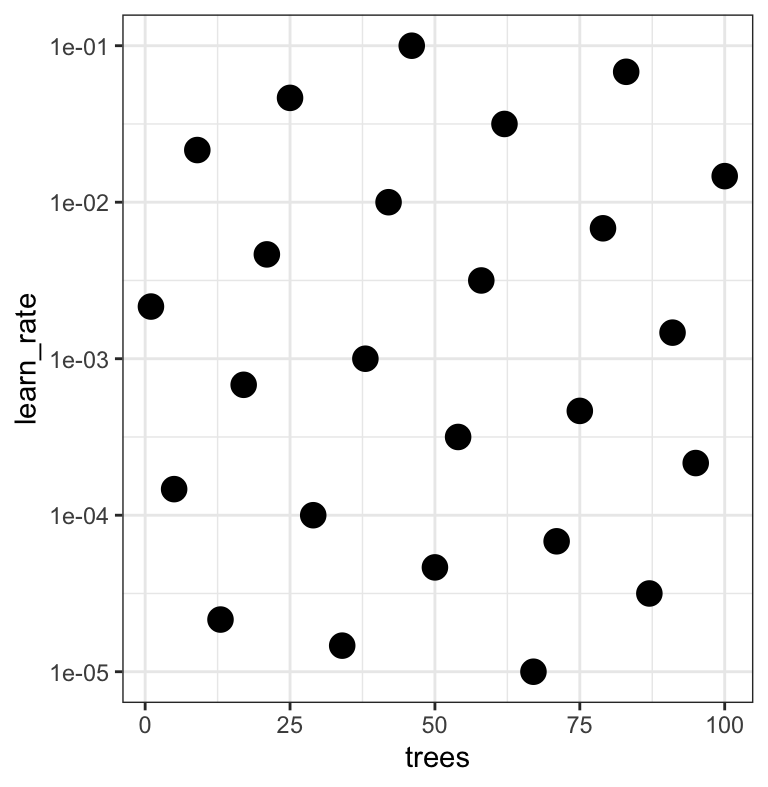
A small grid of points trying to minimize the error via learning rate:
1. Grid search
In reality we would probably sample the space more densely:
2. Iterative Search
We could start with a few points and search the space:
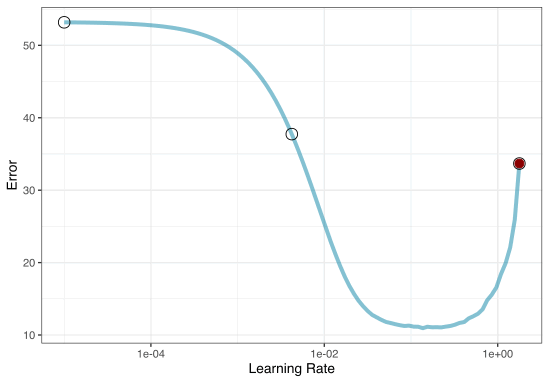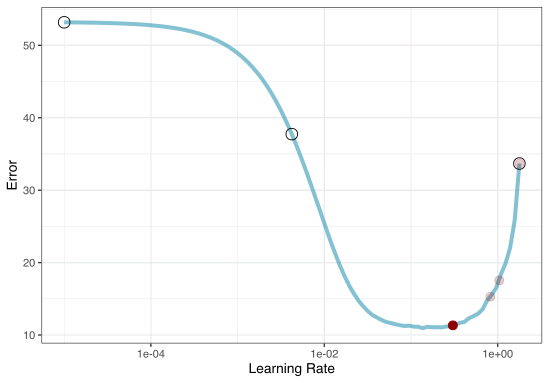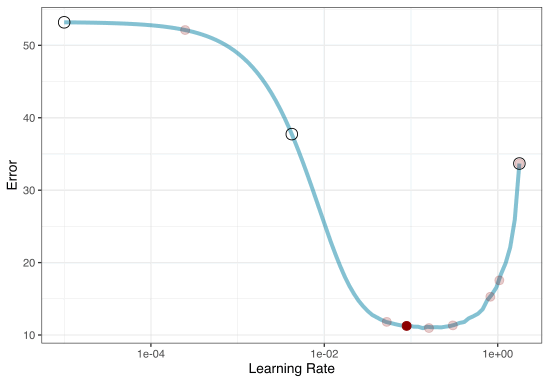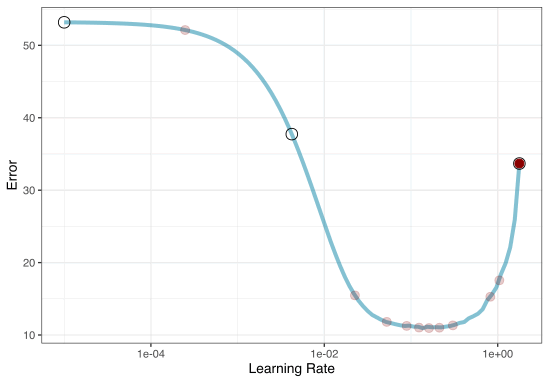
Grid Search Example
Parameters
The tidymodels framework provides pre-defined information on tuning parameters (such as their type, range, transformations, etc).
The
extract_parameter_set_dials()function extracts these tuning parameters and the info.
Grids
Create your grid manually or automatically.
The
grid_*()functions can make a grid.
Different types of grids ![]()
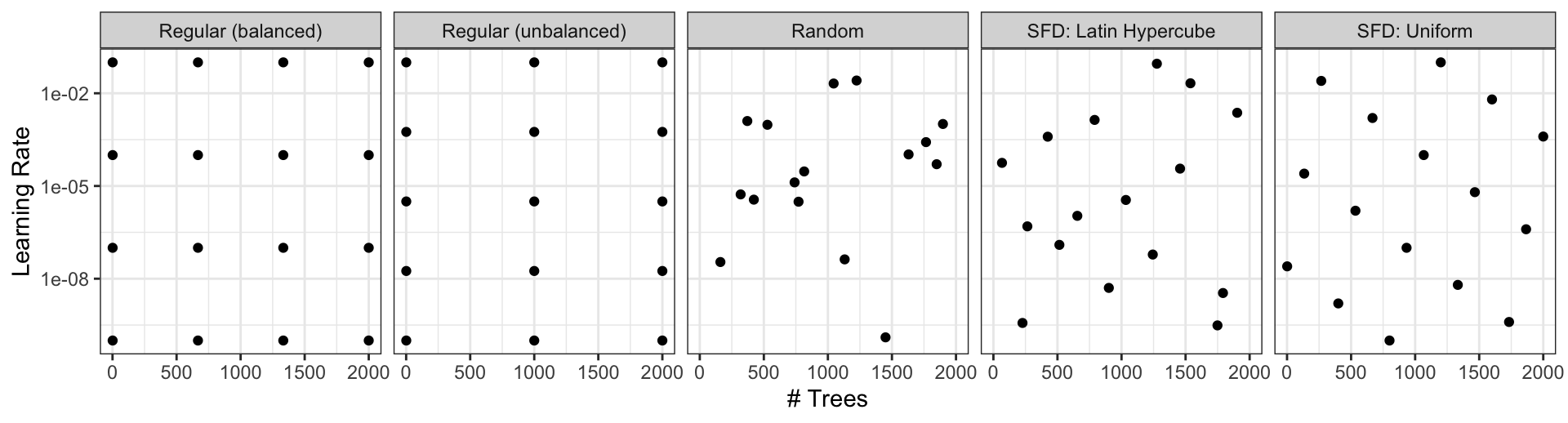
Space-filling designs (SFD) attempt to cover the parameter space without redundant candidates. We recommend these the most.
Create a grid ![]()
![]()
A parameter set can be updated (e.g. to change the ranges).
Create a grid ![]()
![]()
- The
grid_*()functions create a grid of parameter values to evaluate. - The
grid_space_filling()function creates a space-filling design (SFD) of parameter values to evaluate. - The
grid_regular()function creates a regular grid of parameter values to evaluate. - The
grid_random()function creates a random grid of parameter values to evaluate. - The
grid_latin_hypercube()function creates a Latin hypercube design of parameter values to evaluate. - The
grid_max_entropy()function creates a maximum entropy design of parameter values to evaluate.
Create a SFD curve ![]()
![]()
set.seed(12)
(grid <-
b_wflow |>
extract_parameter_set_dials() |>
grid_space_filling(size = 25))
#> # A tibble: 25 × 2
#> trees learn_rate
#> <int> <dbl>
#> 1 1 0.0287
#> 2 84 0.00536
#> 3 167 0.121
#> 4 250 0.00162
#> 5 334 0.0140
#> 6 417 0.0464
#> 7 500 0.196
#> 8 584 0.00422
#> 9 667 0.00127
#> 10 750 0.0178
#> # ℹ 15 more rowsCreate a regular grid ![]()
![]()
set.seed(12)
(grid <-
b_wflow |>
extract_parameter_set_dials() |>
grid_regular(levels = 4))
#> # A tibble: 16 × 2
#> trees learn_rate
#> <int> <dbl>
#> 1 1 0.001
#> 2 667 0.001
#> 3 1333 0.001
#> 4 2000 0.001
#> 5 1 0.00681
#> 6 667 0.00681
#> 7 1333 0.00681
#> 8 2000 0.00681
#> 9 1 0.0464
#> 10 667 0.0464
#> 11 1333 0.0464
#> 12 2000 0.0464
#> 13 1 0.316
#> 14 667 0.316
#> 15 1333 0.316
#> 16 2000 0.316Update parameter ranges ![]()
![]()
b_param <-
b_wflow |>
extract_parameter_set_dials() |>
update(trees = trees(c(1L, 100L)),
learn_rate = learn_rate(c(-5, -1)))
set.seed(712)
(grid <-
b_param |>
grid_space_filling(size = 25))
#> # A tibble: 25 × 2
#> trees learn_rate
#> <int> <dbl>
#> 1 1 0.00215
#> 2 5 0.000147
#> 3 9 0.0215
#> 4 13 0.0000215
#> 5 17 0.000681
#> 6 21 0.00464
#> 7 25 0.0464
#> 8 29 0.0001
#> 9 34 0.0000147
#> 10 38 0.001
#> # ℹ 15 more rowsThe results ![]()
![]()
Note that the learning rates are uniform on the log-10 scale and this shows 2 of 4 dimensions.
Use the tune_*() functions to tune models
Choosing tuning parameters ![]()
![]()
![]()
![]()
Let’s take our previous model and tune more parameters:
Grid Search ![]()
![]()
![]()
Grid Search ![]()
![]()
![]()
b_res
#> # Tuning results
#> # 10-fold cross-validation
#> # A tibble: 10 × 5
#> splits id .metrics .notes .predictions
#> <list> <chr> <list> <list> <list>
#> 1 <split [513/57]> Fold01 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 2 <split [513/57]> Fold02 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 3 <split [513/57]> Fold03 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 4 <split [513/57]> Fold04 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 5 <split [513/57]> Fold05 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 6 <split [513/57]> Fold06 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 7 <split [513/57]> Fold07 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 8 <split [513/57]> Fold08 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 9 <split [513/57]> Fold09 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>
#> 10 <split [513/57]> Fold10 <tibble [25 × 7]> <tibble [0 × 3]> <tibble [1,425 × 7]>Grid results ![]()
Tuning results ![]()
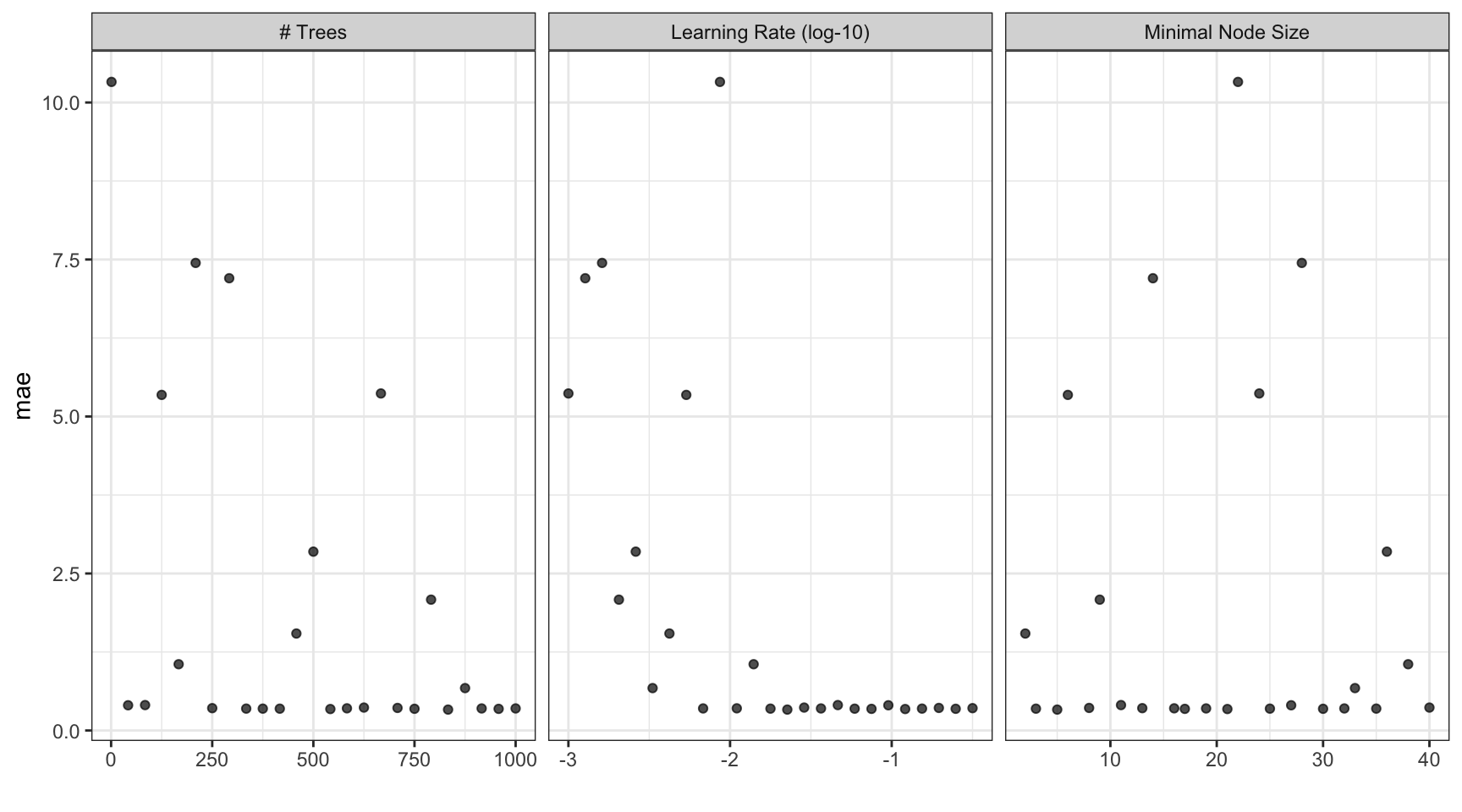
collect_metrics(b_res)
#> # A tibble: 25 × 9
#> trees min_n learn_rate .metric .estimator mean n std_err .config
#> <int> <int> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
#> 1 458 2 0.00422 mae standard 1.54 10 0.0259 Preprocessor1_Model01
#> 2 417 3 0.0590 mae standard 0.347 10 0.0143 Preprocessor1_Model02
#> 3 833 5 0.0226 mae standard 0.333 10 0.0133 Preprocessor1_Model03
#> 4 125 6 0.00536 mae standard 5.34 10 0.0476 Preprocessor1_Model04
#> 5 708 8 0.196 mae standard 0.359 10 0.0120 Preprocessor1_Model05
#> 6 791 9 0.00205 mae standard 2.08 10 0.0300 Preprocessor1_Model06
#> 7 84 11 0.0464 mae standard 0.405 10 0.0186 Preprocessor1_Model07
#> 8 250 13 0.316 mae standard 0.356 10 0.0133 Preprocessor1_Model08
#> 9 292 14 0.00127 mae standard 7.20 10 0.0562 Preprocessor1_Model09
#> 10 583 16 0.0110 mae standard 0.353 10 0.0153 Preprocessor1_Model10
#> # ℹ 15 more rowsChoose a parameter combination ![]()
show_best(b_res, metric = "mae")
#> # A tibble: 5 × 9
#> trees min_n learn_rate .metric .estimator mean n std_err .config
#> <int> <int> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
#> 1 833 5 0.0226 mae standard 0.333 10 0.0133 Preprocessor1_Model03
#> 2 542 21 0.121 mae standard 0.342 10 0.0146 Preprocessor1_Model13
#> 3 958 17 0.0750 mae standard 0.344 10 0.0130 Preprocessor1_Model11
#> 4 750 30 0.249 mae standard 0.346 10 0.0123 Preprocessor1_Model19
#> 5 417 3 0.0590 mae standard 0.347 10 0.0143 Preprocessor1_Model02Choose a parameter combination ![]()
Create your own tibble for final parameters or use one of the tune::select_*() functions:
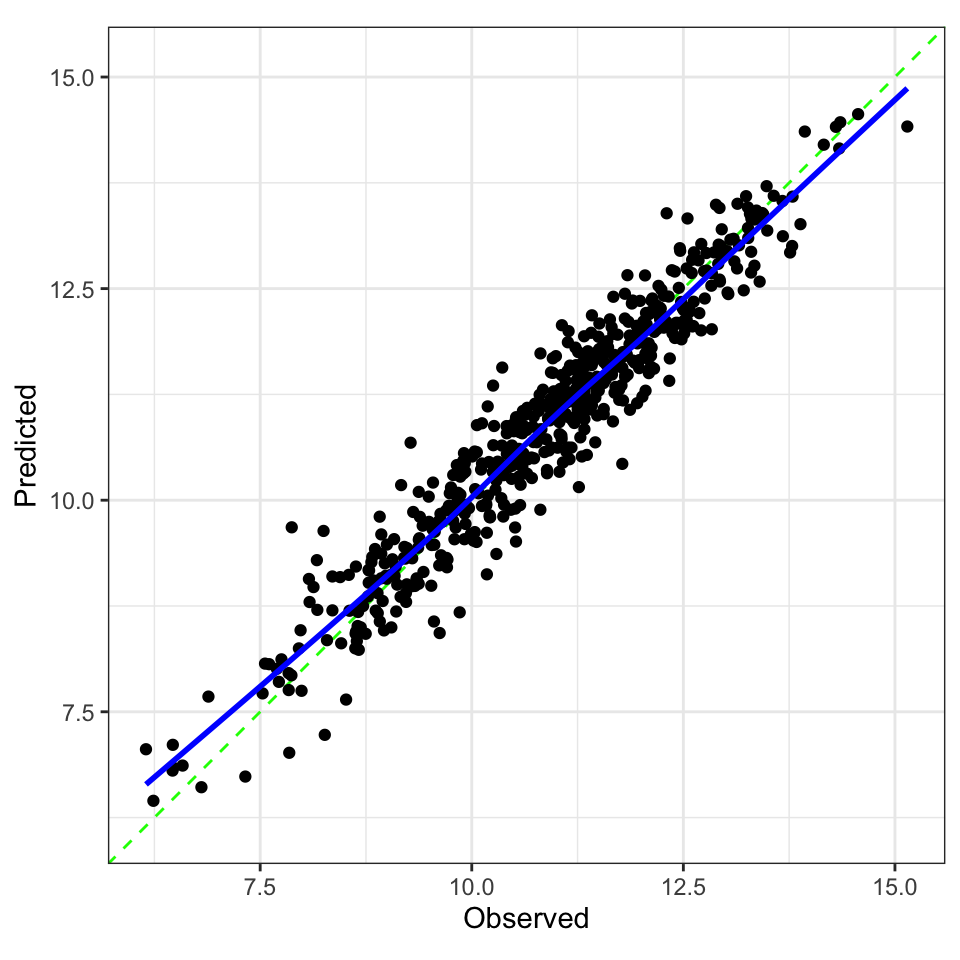
Checking Calibration ![]()
![]()
The final fit!
Suppose that we are happy with our model.
Let’s fit the model on the training set and verify our performance using the test set.
We’ve seen fit() and predict() (+ augment()) but there is a shortcut:
# the boosted tree workflow can be `finalized` with the best parameters
workflow <- finalize_workflow(b_wflow, b_best)
# forested_split has train + test info
(final_fit <- last_fit(workflow, split))
#> # Resampling results
#> # Manual resampling
#> # A tibble: 1 × 6
#> splits id .metrics .notes .predictions .workflow
#> <list> <chr> <list> <list> <list> <list>
#> 1 <split [570/144]> train/test split <tibble [2 × 4]> <tibble [0 × 3]> <tibble [144 × 4]> <workflow>The final fit!
collect_metrics(final_fit)
#> # A tibble: 2 × 4
#> .metric .estimator .estimate .config
#> <chr> <chr> <dbl> <chr>
#> 1 rmse standard 0.391 Preprocessor1_Model1
#> 2 rsq standard 0.918 Preprocessor1_Model1
collect_predictions(final_fit) |>
ggplot(aes(.pred, logC)) +
geom_point() +
geom_abline() +
geom_smooth(method = "lm", se = FALSE)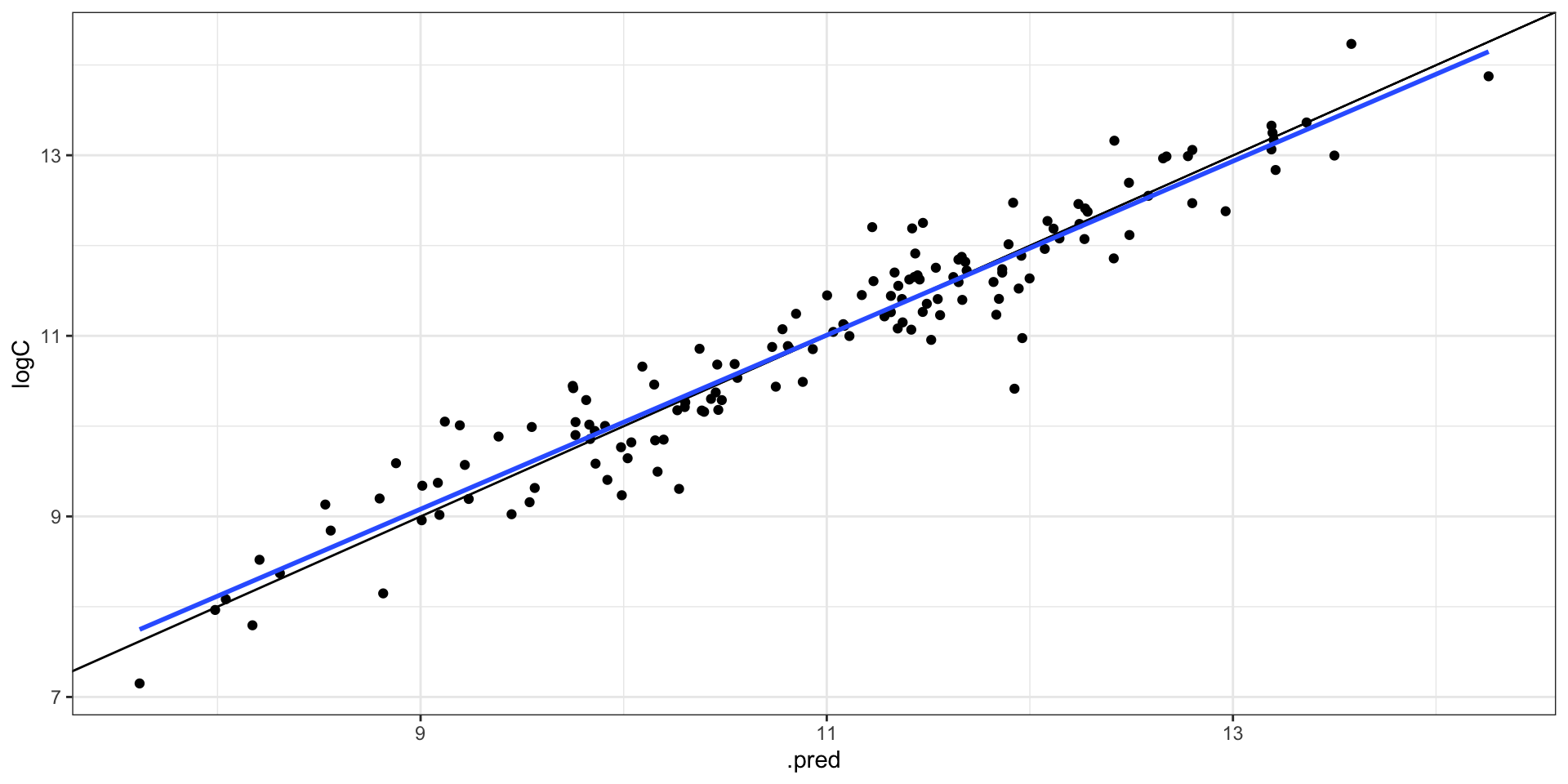
The whole game
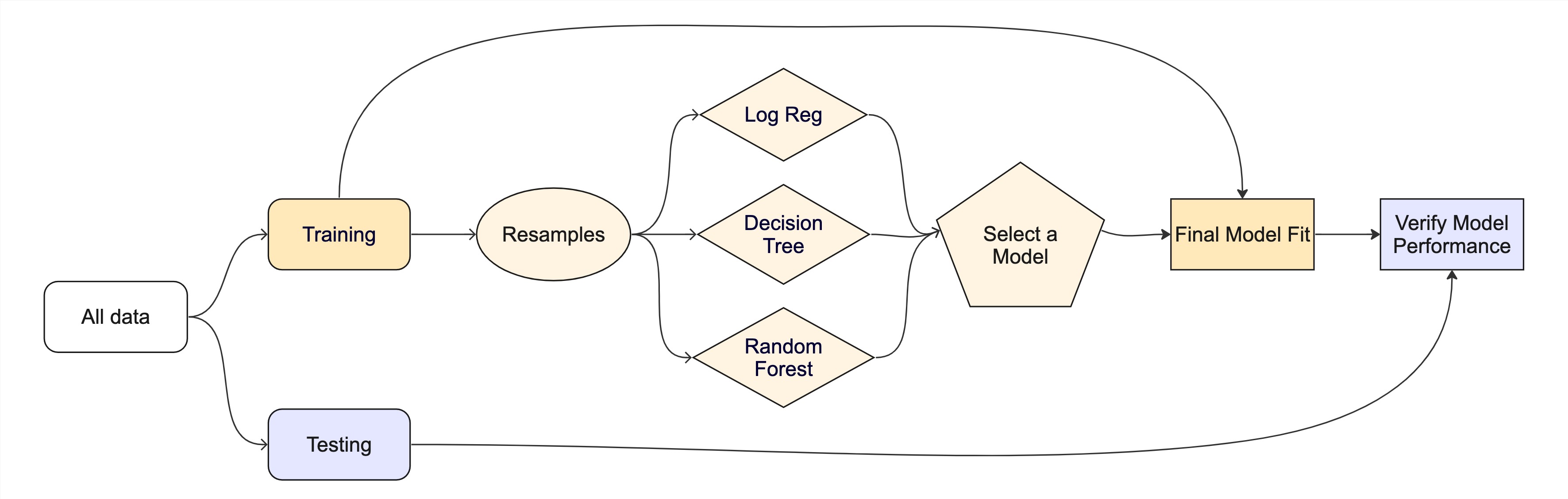
Assignment
Starting with the “Whole Game Image” on the last slide, in plain language, write a sentence or 2 about each step and item, its importance, and note key things to be careful of. The full submission should be a 2-3 paragraph narative of the entire process, including the final model fit and performance.