library(tidyverse)
url <- 'https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-counties.csv'
covid <- read_csv(url)
head(covid)
# A tibble: 6 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-21 Snohomish Washington 53061 1 0
2 2020-01-22 Snohomish Washington 53061 1 0
3 2020-01-23 Snohomish Washington 53061 1 0
4 2020-01-24 Cook Illinois 17031 1 0
5 2020-01-24 Snohomish Washington 53061 1 0
6 2020-01-25 Orange California 06059 1 0Lecture 08
Relations & Data Format
2025-02-23
Picking back up!
We’ve covered many topics on how to manipulate and reshape a single data.frame:
Storage vs Implementation: Bytes/Bytes vs Interpretation
Location: remote/local/memory
Data: types & structures
Manipulation: dplyr verbs: grammar of data manipulation
Visualization: ggplot2: grammar of data visualization
Today: When one table is not enough, when tidy isn’t “right”
Yesterdays Assignment
Make a faceted line plot (geom_line) of the 6 states with most cases. Your X axis should be the date and the y axis cases.
Make a column plot (
geom_col) of daily total cases in the USA. Your X axis should be the date and the y axis cases.
Question 1: Close…
# A tibble: 2,502,832 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-21 Snohomish Washington 53061 1 0
2 2020-01-22 Snohomish Washington 53061 1 0
3 2020-01-23 Snohomish Washington 53061 1 0
4 2020-01-24 Cook Illinois 17031 1 0
5 2020-01-24 Snohomish Washington 53061 1 0
6 2020-01-25 Orange California 06059 1 0
7 2020-01-25 Cook Illinois 17031 1 0
8 2020-01-25 Snohomish Washington 53061 1 0
9 2020-01-26 Maricopa Arizona 04013 1 0
10 2020-01-26 Los Angeles California 06037 1 0
# ℹ 2,502,822 more rowsQuestion 1: Close…
# A tibble: 3,258 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2022-05-13 Autauga Alabama 01001 15863 216
2 2022-05-13 Baldwin Alabama 01003 55862 681
3 2022-05-13 Barbour Alabama 01005 5681 98
4 2022-05-13 Bibb Alabama 01007 6457 105
5 2022-05-13 Blount Alabama 01009 15005 243
6 2022-05-13 Bullock Alabama 01011 2319 54
7 2022-05-13 Butler Alabama 01013 5068 129
8 2022-05-13 Calhoun Alabama 01015 32453 627
9 2022-05-13 Chambers Alabama 01017 8508 162
10 2022-05-13 Cherokee Alabama 01019 5131 86
# ℹ 3,248 more rowsQuestion 1: Close…
# A tibble: 3,258 × 6
# Groups: state [56]
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2022-05-13 Autauga Alabama 01001 15863 216
2 2022-05-13 Baldwin Alabama 01003 55862 681
3 2022-05-13 Barbour Alabama 01005 5681 98
4 2022-05-13 Bibb Alabama 01007 6457 105
5 2022-05-13 Blount Alabama 01009 15005 243
6 2022-05-13 Bullock Alabama 01011 2319 54
7 2022-05-13 Butler Alabama 01013 5068 129
8 2022-05-13 Calhoun Alabama 01015 32453 627
9 2022-05-13 Chambers Alabama 01017 8508 162
10 2022-05-13 Cherokee Alabama 01019 5131 86
# ℹ 3,248 more rowsQuestion 1: Close…
# A tibble: 56 × 2
state cases
<chr> <dbl>
1 Alabama 1304710
2 Alaska 254467
3 American Samoa 5930
4 Arizona 2030925
5 Arkansas 838251
6 California 9351630
7 Colorado 1412121
8 Connecticut 779460
9 Delaware 267265
10 District of Columbia 143943
# ℹ 46 more rowsQuestion 1: Close…
# A tibble: 56 × 2
state cases
<chr> <dbl>
1 Alabama 1304710
2 Alaska 254467
3 American Samoa 5930
4 Arizona 2030925
5 Arkansas 838251
6 California 9351630
7 Colorado 1412121
8 Connecticut 779460
9 Delaware 267265
10 District of Columbia 143943
# ℹ 46 more rowsQuestion 1: Close…
Question 1: Close…
Question 1: Close…
[1] "California" "Texas" "Florida" "New York" "Illinois"
[6] "Pennsylvania"
# A tibble: 2,502,832 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-21 Snohomish Washington 53061 1 0
2 2020-01-22 Snohomish Washington 53061 1 0
3 2020-01-23 Snohomish Washington 53061 1 0
4 2020-01-24 Cook Illinois 17031 1 0
5 2020-01-24 Snohomish Washington 53061 1 0
6 2020-01-25 Orange California 06059 1 0
7 2020-01-25 Cook Illinois 17031 1 0
8 2020-01-25 Snohomish Washington 53061 1 0
9 2020-01-26 Maricopa Arizona 04013 1 0
10 2020-01-26 Los Angeles California 06037 1 0
# ℹ 2,502,822 more rowsQuestion 1: Close…
[1] "California" "Texas" "Florida" "New York" "Illinois"
[6] "Pennsylvania"
# A tibble: 470,209 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-24 Cook Illinois 17031 1 0
2 2020-01-25 Orange California 06059 1 0
3 2020-01-25 Cook Illinois 17031 1 0
4 2020-01-26 Los Angeles California 06037 1 0
5 2020-01-26 Orange California 06059 1 0
6 2020-01-26 Cook Illinois 17031 1 0
7 2020-01-27 Los Angeles California 06037 1 0
8 2020-01-27 Orange California 06059 1 0
9 2020-01-27 Cook Illinois 17031 1 0
10 2020-01-28 Los Angeles California 06037 1 0
# ℹ 470,199 more rowsQuestion 1: Close…
[1] "California" "Texas" "Florida" "New York" "Illinois"
[6] "Pennsylvania"
Question 1: Close…
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state)
covid |>
filter(state %in% c("California", "Texas", "Florida", "New York", "Illinois", "Pennsylvania")) |>
ggplot(aes(x = date, y = cases)) +
geom_line(aes(color = state))[1] "California" "Texas" "Florida" "New York" "Illinois"
[6] "Pennsylvania"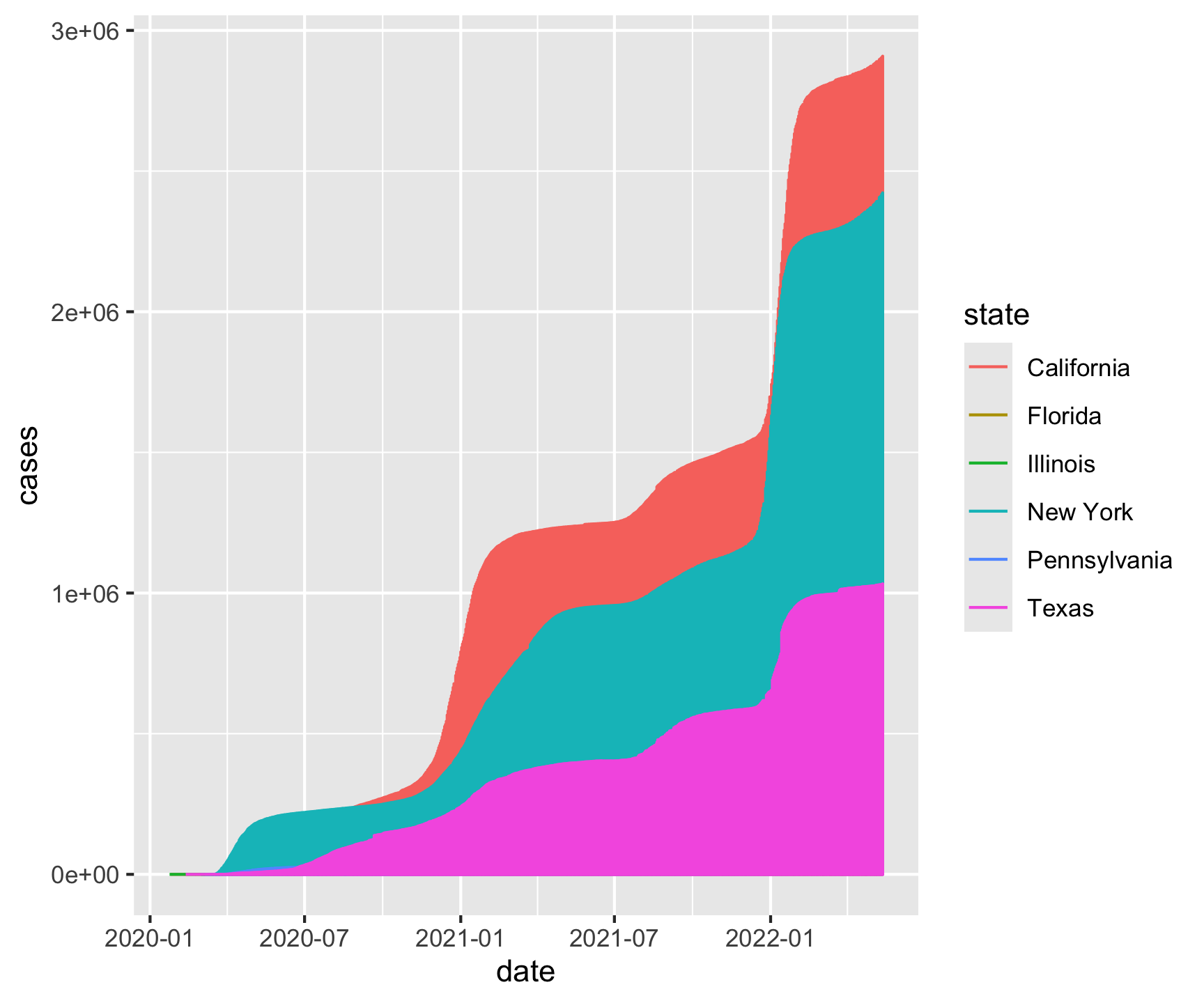
Question 1: Close…
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state)
covid |>
filter(state %in% c("California", "Texas", "Florida", "New York", "Illinois", "Pennsylvania")) |>
ggplot(aes(x = date, y = cases)) +
geom_line(aes(color = state)) +
facet_wrap(~state)[1] "California" "Texas" "Florida" "New York" "Illinois"
[6] "Pennsylvania"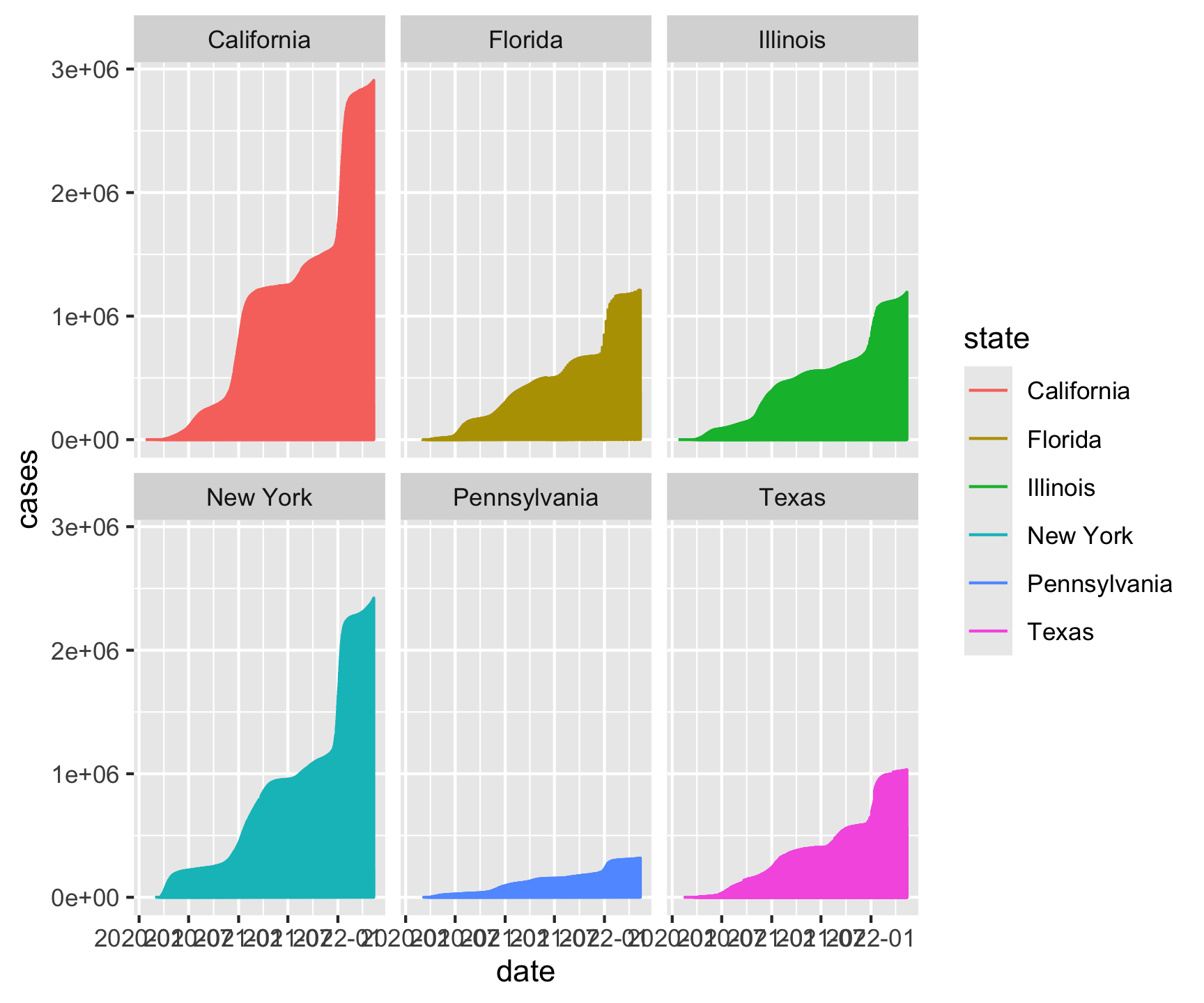
Question 1: Close…
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state)
covid |>
filter(state %in% c("California", "Texas", "Florida", "New York", "Illinois", "Pennsylvania")) |>
ggplot(aes(x = date, y = cases)) +
geom_line(aes(color = state)) +
facet_wrap(~state) +
theme_gray()[1] "California" "Texas" "Florida" "New York" "Illinois"
[6] "Pennsylvania"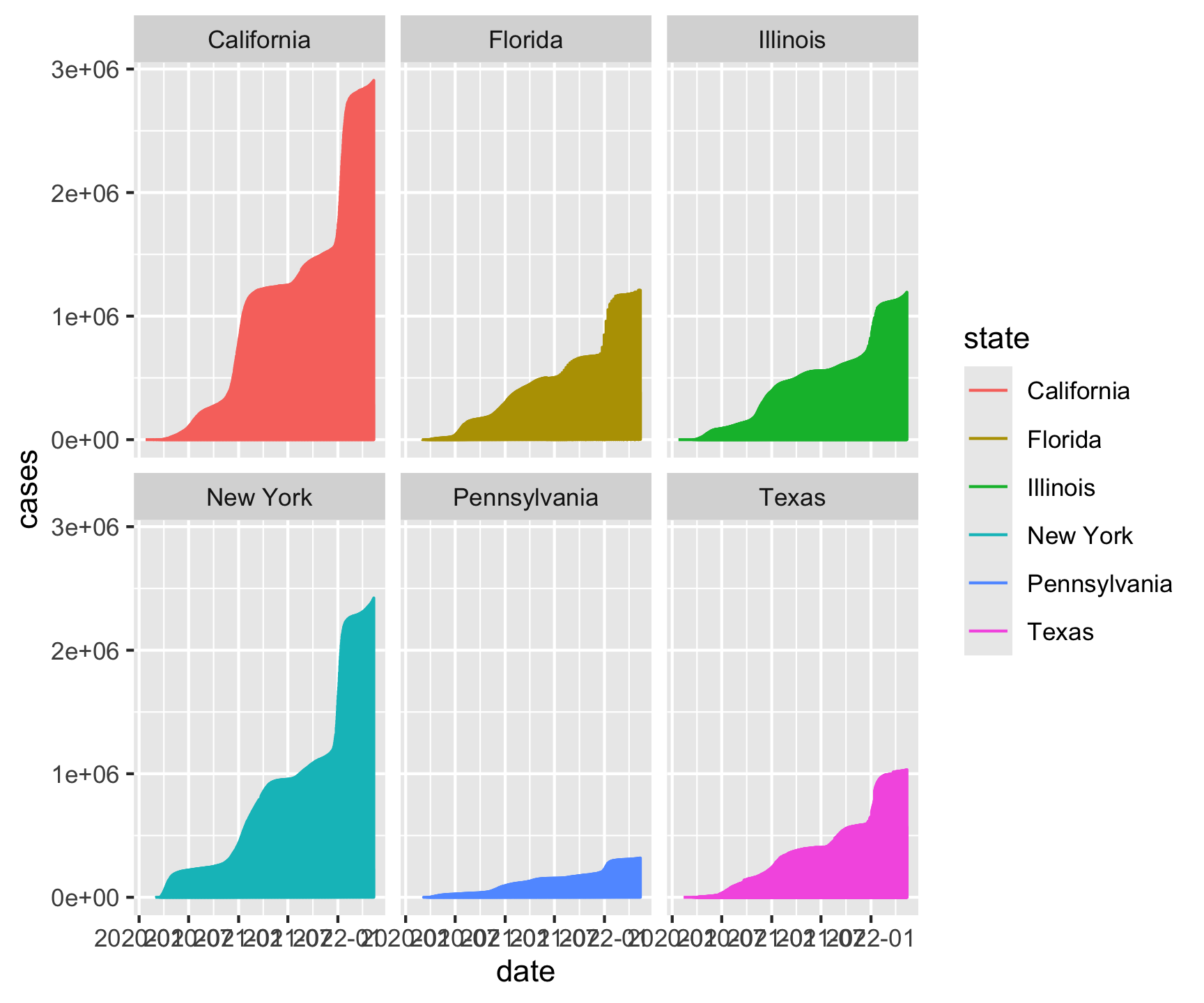
Question 1: State Level
# A tibble: 2,502,832 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-21 Snohomish Washington 53061 1 0
2 2020-01-22 Snohomish Washington 53061 1 0
3 2020-01-23 Snohomish Washington 53061 1 0
4 2020-01-24 Cook Illinois 17031 1 0
5 2020-01-24 Snohomish Washington 53061 1 0
6 2020-01-25 Orange California 06059 1 0
7 2020-01-25 Cook Illinois 17031 1 0
8 2020-01-25 Snohomish Washington 53061 1 0
9 2020-01-26 Maricopa Arizona 04013 1 0
10 2020-01-26 Los Angeles California 06037 1 0
# ℹ 2,502,822 more rowsQuestion 1: State Level
# A tibble: 3,258 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2022-05-13 Autauga Alabama 01001 15863 216
2 2022-05-13 Baldwin Alabama 01003 55862 681
3 2022-05-13 Barbour Alabama 01005 5681 98
4 2022-05-13 Bibb Alabama 01007 6457 105
5 2022-05-13 Blount Alabama 01009 15005 243
6 2022-05-13 Bullock Alabama 01011 2319 54
7 2022-05-13 Butler Alabama 01013 5068 129
8 2022-05-13 Calhoun Alabama 01015 32453 627
9 2022-05-13 Chambers Alabama 01017 8508 162
10 2022-05-13 Cherokee Alabama 01019 5131 86
# ℹ 3,248 more rowsQuestion 1: State Level
# A tibble: 3,258 × 6
# Groups: state [56]
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2022-05-13 Autauga Alabama 01001 15863 216
2 2022-05-13 Baldwin Alabama 01003 55862 681
3 2022-05-13 Barbour Alabama 01005 5681 98
4 2022-05-13 Bibb Alabama 01007 6457 105
5 2022-05-13 Blount Alabama 01009 15005 243
6 2022-05-13 Bullock Alabama 01011 2319 54
7 2022-05-13 Butler Alabama 01013 5068 129
8 2022-05-13 Calhoun Alabama 01015 32453 627
9 2022-05-13 Chambers Alabama 01017 8508 162
10 2022-05-13 Cherokee Alabama 01019 5131 86
# ℹ 3,248 more rowsQuestion 1: State Level
# A tibble: 56 × 2
state cases
<chr> <dbl>
1 Alabama 1304710
2 Alaska 254467
3 American Samoa 5930
4 Arizona 2030925
5 Arkansas 838251
6 California 9351630
7 Colorado 1412121
8 Connecticut 779460
9 Delaware 267265
10 District of Columbia 143943
# ℹ 46 more rowsQuestion 1: State Level
# A tibble: 56 × 2
state cases
<chr> <dbl>
1 Alabama 1304710
2 Alaska 254467
3 American Samoa 5930
4 Arizona 2030925
5 Arkansas 838251
6 California 9351630
7 Colorado 1412121
8 Connecticut 779460
9 Delaware 267265
10 District of Columbia 143943
# ℹ 46 more rowsQuestion 1: State Level
Question 1: State Level
Question 1: State Level
Question 1: State Level
# A tibble: 2,502,832 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-21 Snohomish Washington 53061 1 0
2 2020-01-22 Snohomish Washington 53061 1 0
3 2020-01-23 Snohomish Washington 53061 1 0
4 2020-01-24 Cook Illinois 17031 1 0
5 2020-01-24 Snohomish Washington 53061 1 0
6 2020-01-25 Orange California 06059 1 0
7 2020-01-25 Cook Illinois 17031 1 0
8 2020-01-25 Snohomish Washington 53061 1 0
9 2020-01-26 Maricopa Arizona 04013 1 0
10 2020-01-26 Los Angeles California 06037 1 0
# ℹ 2,502,822 more rowsQuestion 1: State Level
# A tibble: 470,209 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-24 Cook Illinois 17031 1 0
2 2020-01-25 Orange California 06059 1 0
3 2020-01-25 Cook Illinois 17031 1 0
4 2020-01-26 Los Angeles California 06037 1 0
5 2020-01-26 Orange California 06059 1 0
6 2020-01-26 Cook Illinois 17031 1 0
7 2020-01-27 Los Angeles California 06037 1 0
8 2020-01-27 Orange California 06059 1 0
9 2020-01-27 Cook Illinois 17031 1 0
10 2020-01-28 Los Angeles California 06037 1 0
# ℹ 470,199 more rowsQuestion 1: State Level
# A tibble: 470,209 × 6
# Groups: state, date [4,910]
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-24 Cook Illinois 17031 1 0
2 2020-01-25 Orange California 06059 1 0
3 2020-01-25 Cook Illinois 17031 1 0
4 2020-01-26 Los Angeles California 06037 1 0
5 2020-01-26 Orange California 06059 1 0
6 2020-01-26 Cook Illinois 17031 1 0
7 2020-01-27 Los Angeles California 06037 1 0
8 2020-01-27 Orange California 06059 1 0
9 2020-01-27 Cook Illinois 17031 1 0
10 2020-01-28 Los Angeles California 06037 1 0
# ℹ 470,199 more rowsQuestion 1: State Level
# A tibble: 4,910 × 3
# Groups: state [6]
state date cases
<chr> <date> <dbl>
1 California 2020-01-25 1
2 California 2020-01-26 2
3 California 2020-01-27 2
4 California 2020-01-28 2
5 California 2020-01-29 2
6 California 2020-01-30 2
7 California 2020-01-31 3
8 California 2020-02-01 3
9 California 2020-02-02 6
10 California 2020-02-03 6
# ℹ 4,900 more rowsQuestion 1: State Level
# A tibble: 4,910 × 3
state date cases
<chr> <date> <dbl>
1 California 2020-01-25 1
2 California 2020-01-26 2
3 California 2020-01-27 2
4 California 2020-01-28 2
5 California 2020-01-29 2
6 California 2020-01-30 2
7 California 2020-01-31 3
8 California 2020-02-01 3
9 California 2020-02-02 6
10 California 2020-02-03 6
# ℹ 4,900 more rowsQuestion 1: State Level
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state) ->
top_states
covid |>
filter(state %in% top_states) |>
group_by(state, date) |>
summarise(cases = sum(cases)) |>
ungroup() |>
ggplot(aes(x = date, y = cases, color = state))
Question 1: State Level
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state) ->
top_states
covid |>
filter(state %in% top_states) |>
group_by(state, date) |>
summarise(cases = sum(cases)) |>
ungroup() |>
ggplot(aes(x = date, y = cases, color = state)) +
geom_line(size = 2)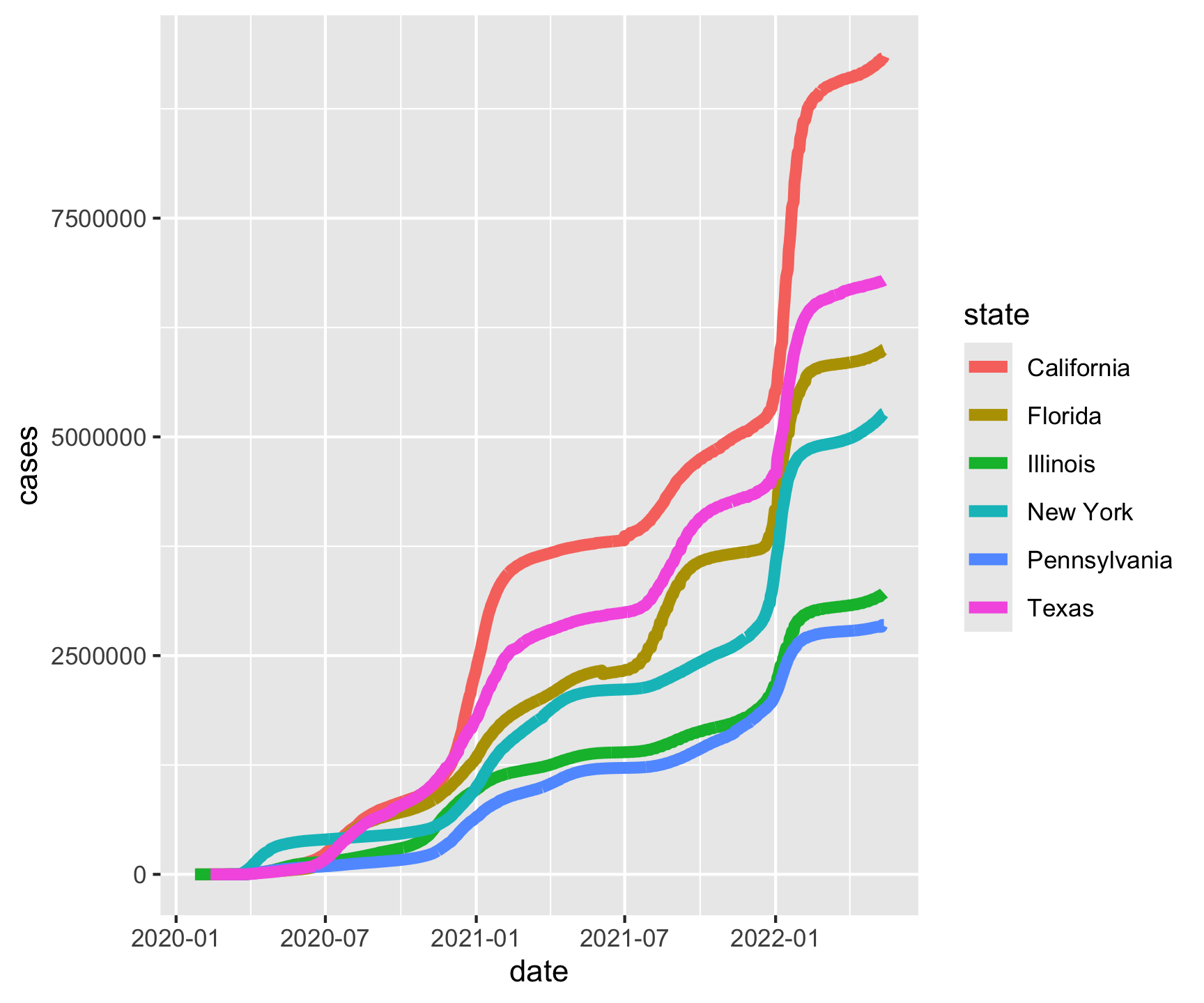
Question 1: State Level
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state) ->
top_states
covid |>
filter(state %in% top_states) |>
group_by(state, date) |>
summarise(cases = sum(cases)) |>
ungroup() |>
ggplot(aes(x = date, y = cases, color = state)) +
geom_line(size = 2) +
facet_wrap(~state)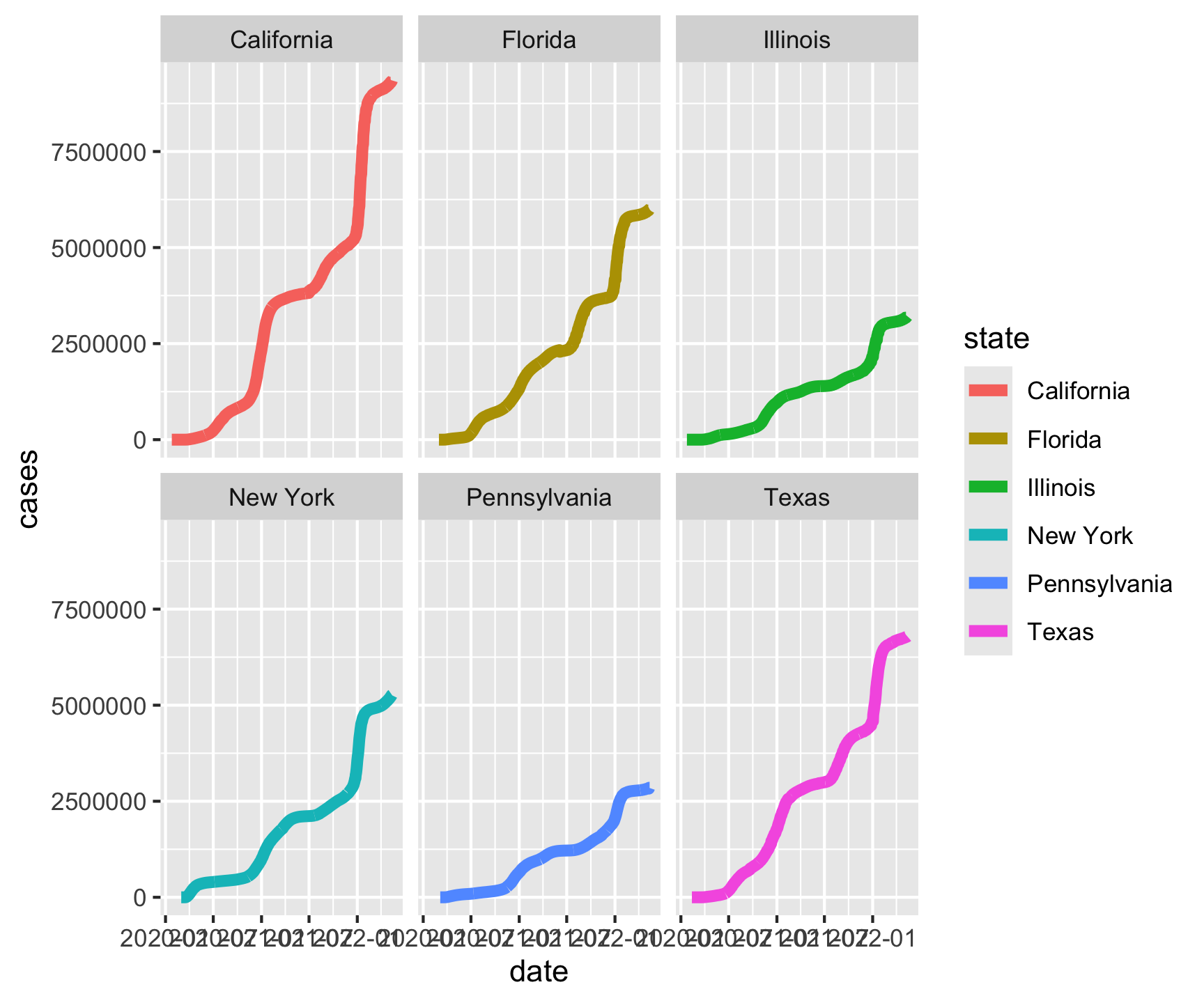
Question 1: State Level
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state) ->
top_states
covid |>
filter(state %in% top_states) |>
group_by(state, date) |>
summarise(cases = sum(cases)) |>
ungroup() |>
ggplot(aes(x = date, y = cases, color = state)) +
geom_line(size = 2) +
facet_wrap(~state) +
ggthemes::theme_gdocs()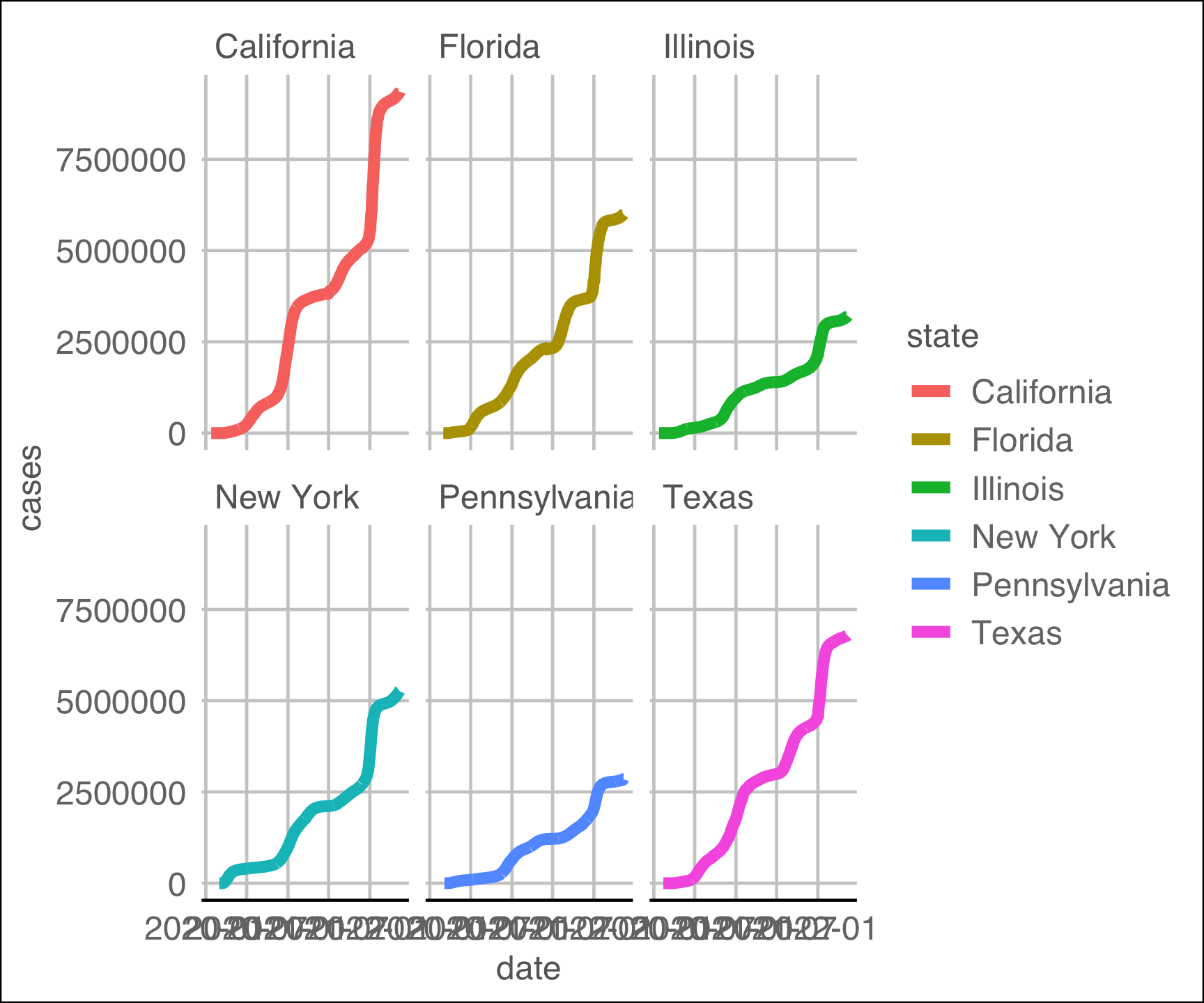
Question 1: State Level
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state) ->
top_states
covid |>
filter(state %in% top_states) |>
group_by(state, date) |>
summarise(cases = sum(cases)) |>
ungroup() |>
ggplot(aes(x = date, y = cases, color = state)) +
geom_line(size = 2) +
facet_wrap(~state) +
ggthemes::theme_gdocs() +
theme(legend.position = 'NA')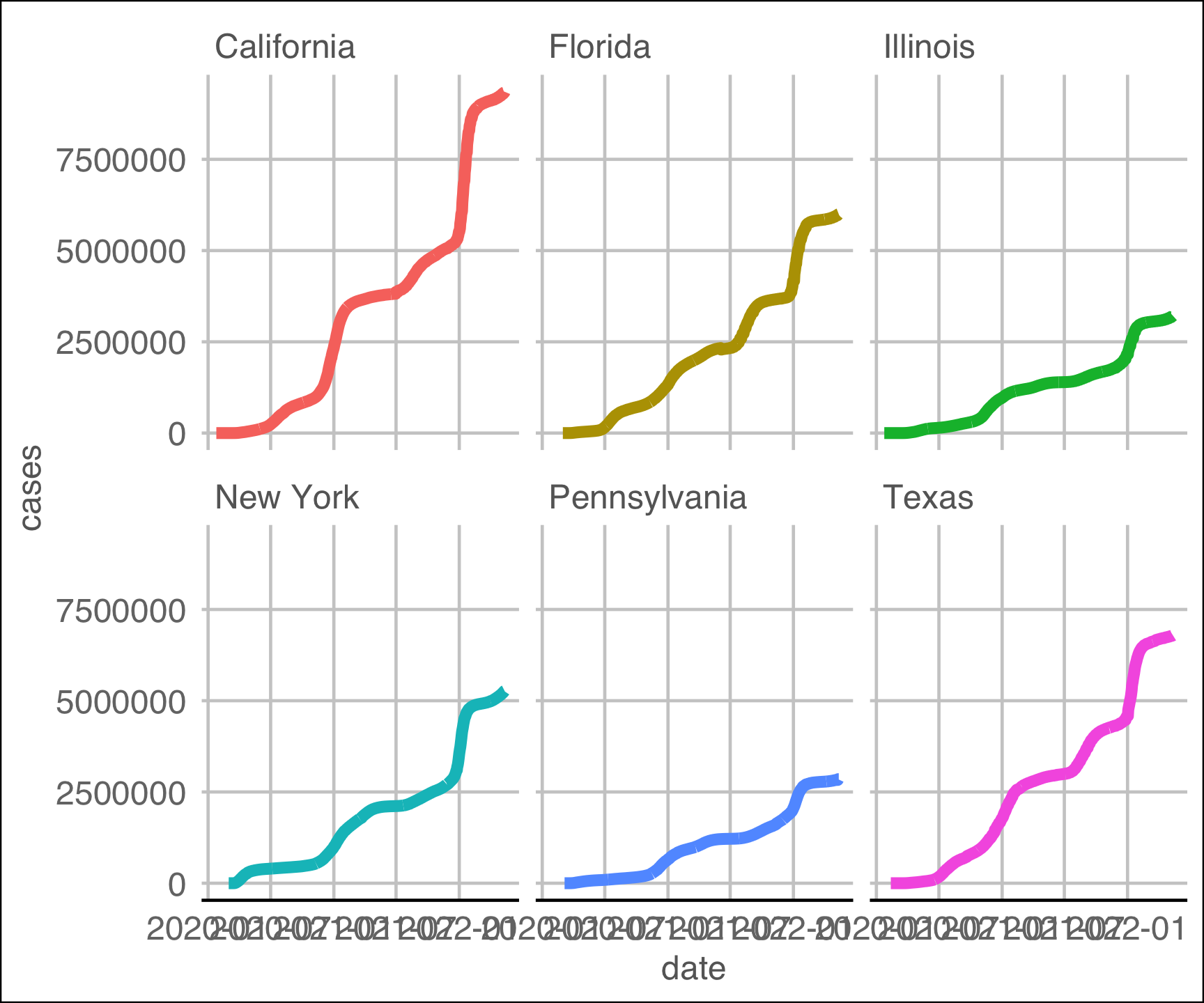
Question 1: State Level
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
slice_max(cases, n = 6) |>
pull(state) ->
top_states
covid |>
filter(state %in% top_states) |>
group_by(state, date) |>
summarise(cases = sum(cases)) |>
ungroup() |>
ggplot(aes(x = date, y = cases, color = state)) +
geom_line(size = 2) +
facet_wrap(~state) +
ggthemes::theme_gdocs() +
theme(legend.position = 'NA') +
labs(title = "Cummulative Case Counts",
subtitle = "Data Source: NY-Times",
x = "Date",
y = "Cases",
caption = "Daily Exercise 07")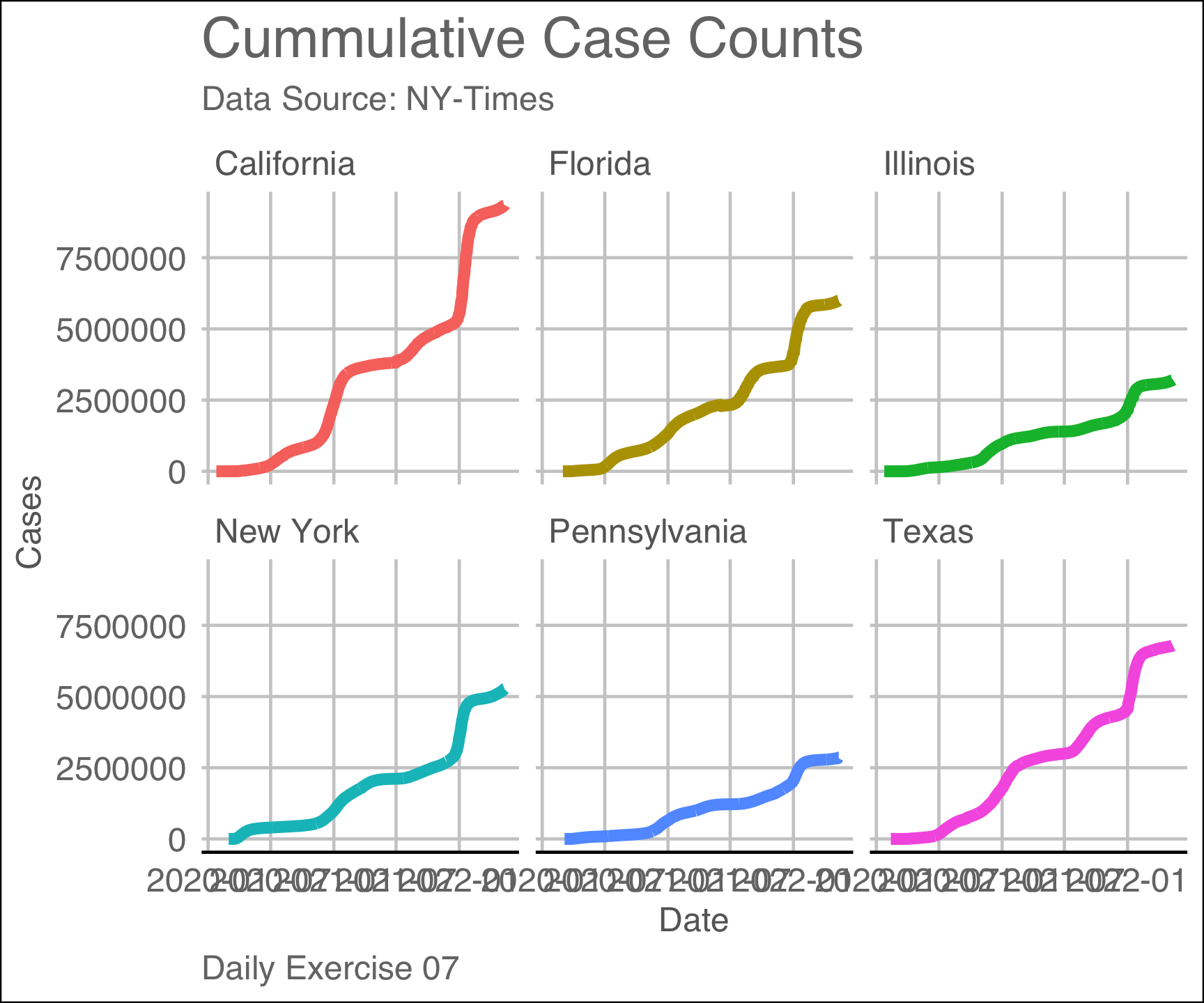
Question 2: National Level
# A tibble: 2,502,832 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-21 Snohomish Washington 53061 1 0
2 2020-01-22 Snohomish Washington 53061 1 0
3 2020-01-23 Snohomish Washington 53061 1 0
4 2020-01-24 Cook Illinois 17031 1 0
5 2020-01-24 Snohomish Washington 53061 1 0
6 2020-01-25 Orange California 06059 1 0
7 2020-01-25 Cook Illinois 17031 1 0
8 2020-01-25 Snohomish Washington 53061 1 0
9 2020-01-26 Maricopa Arizona 04013 1 0
10 2020-01-26 Los Angeles California 06037 1 0
# ℹ 2,502,822 more rowsQuestion 2: National Level
# A tibble: 2,502,832 × 6
# Groups: date [844]
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-21 Snohomish Washington 53061 1 0
2 2020-01-22 Snohomish Washington 53061 1 0
3 2020-01-23 Snohomish Washington 53061 1 0
4 2020-01-24 Cook Illinois 17031 1 0
5 2020-01-24 Snohomish Washington 53061 1 0
6 2020-01-25 Orange California 06059 1 0
7 2020-01-25 Cook Illinois 17031 1 0
8 2020-01-25 Snohomish Washington 53061 1 0
9 2020-01-26 Maricopa Arizona 04013 1 0
10 2020-01-26 Los Angeles California 06037 1 0
# ℹ 2,502,822 more rowsQuestion 2: National Level
Question 2: National Level
Question 2: National Level
Question 2: National Level
Question 2: National Level
Question 2: National Level
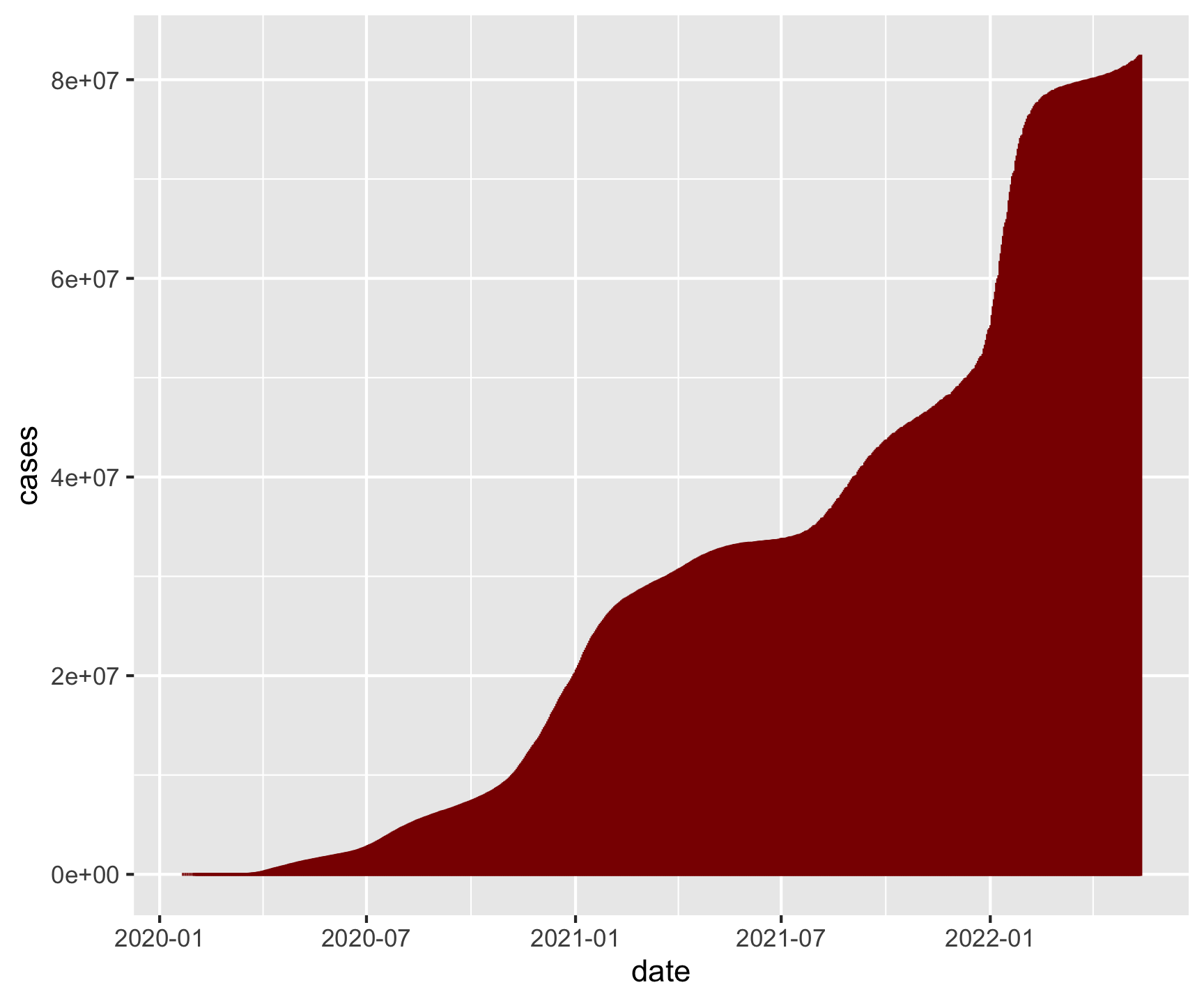
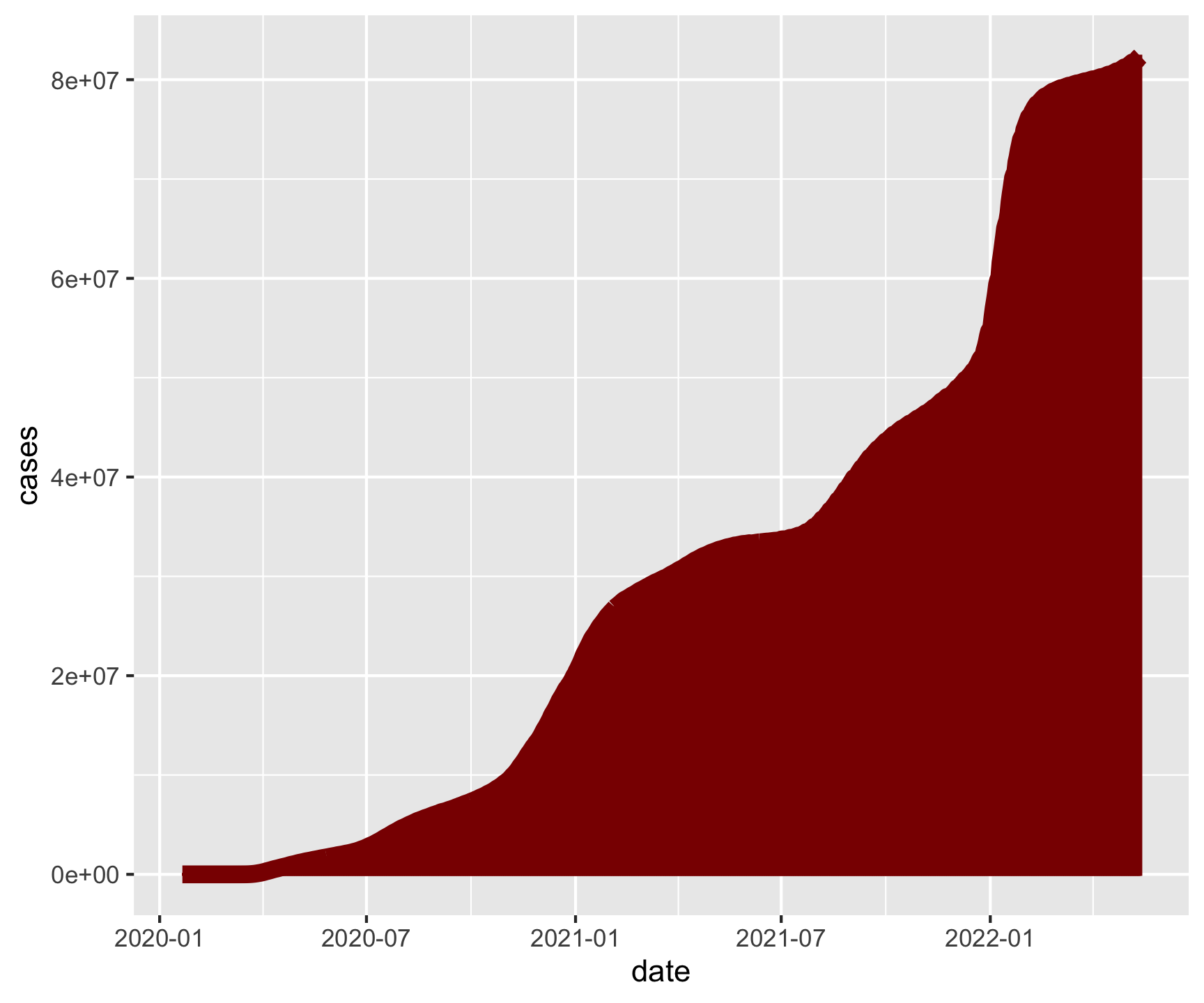
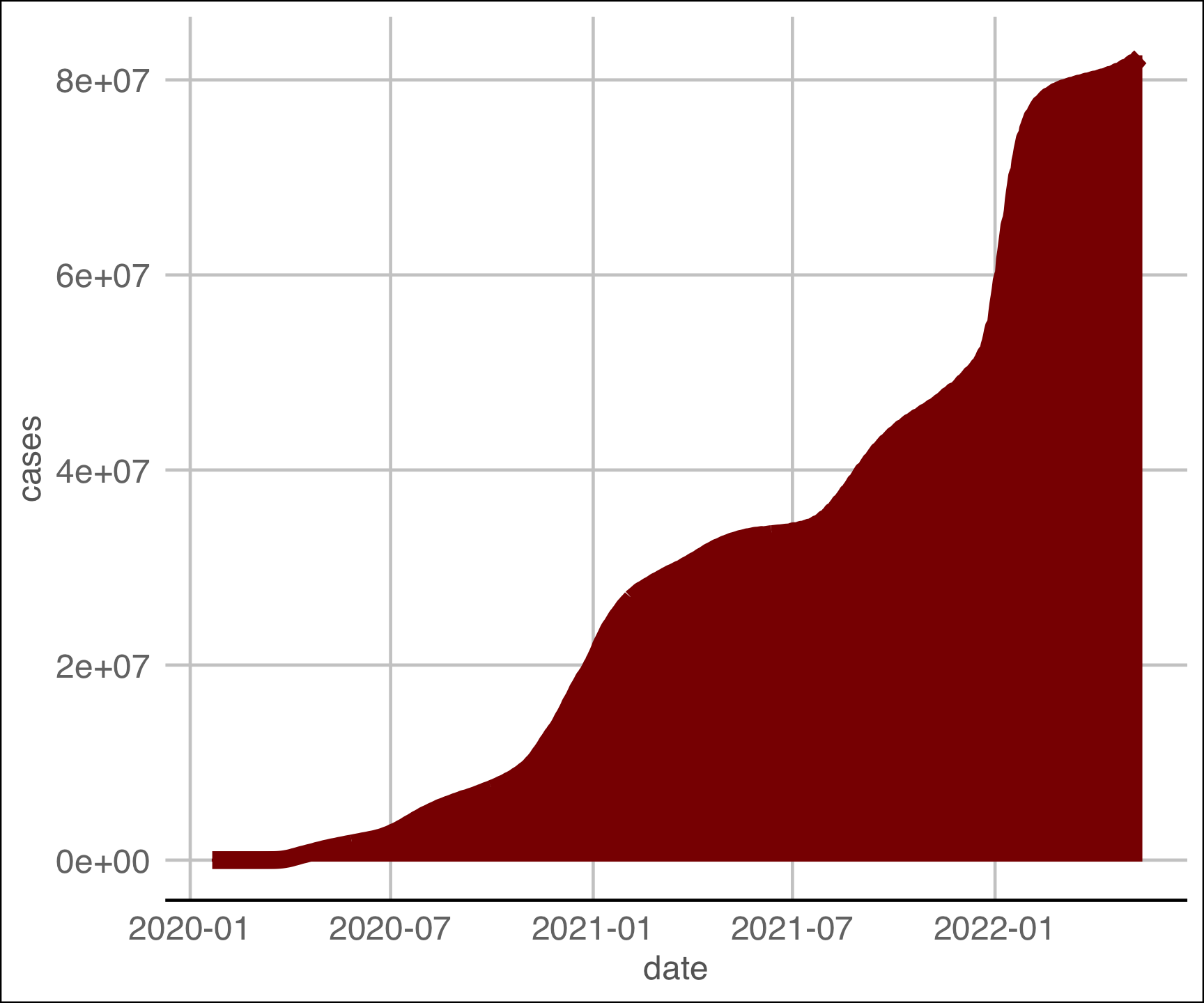
covid |>
group_by(date) |>
summarize(cases = sum(cases)) |>
ggplot(aes(x = date, y = cases)) +
geom_col(fill = "darkred", color = "darkred", alpha = .25) +
geom_line(color = "darkred", size = 3) +
ggthemes::theme_gdocs() +
labs(title = "National Cummulative Case Counts",
x = "Date",
y = "Cases",
caption = "Daily Exercise 07")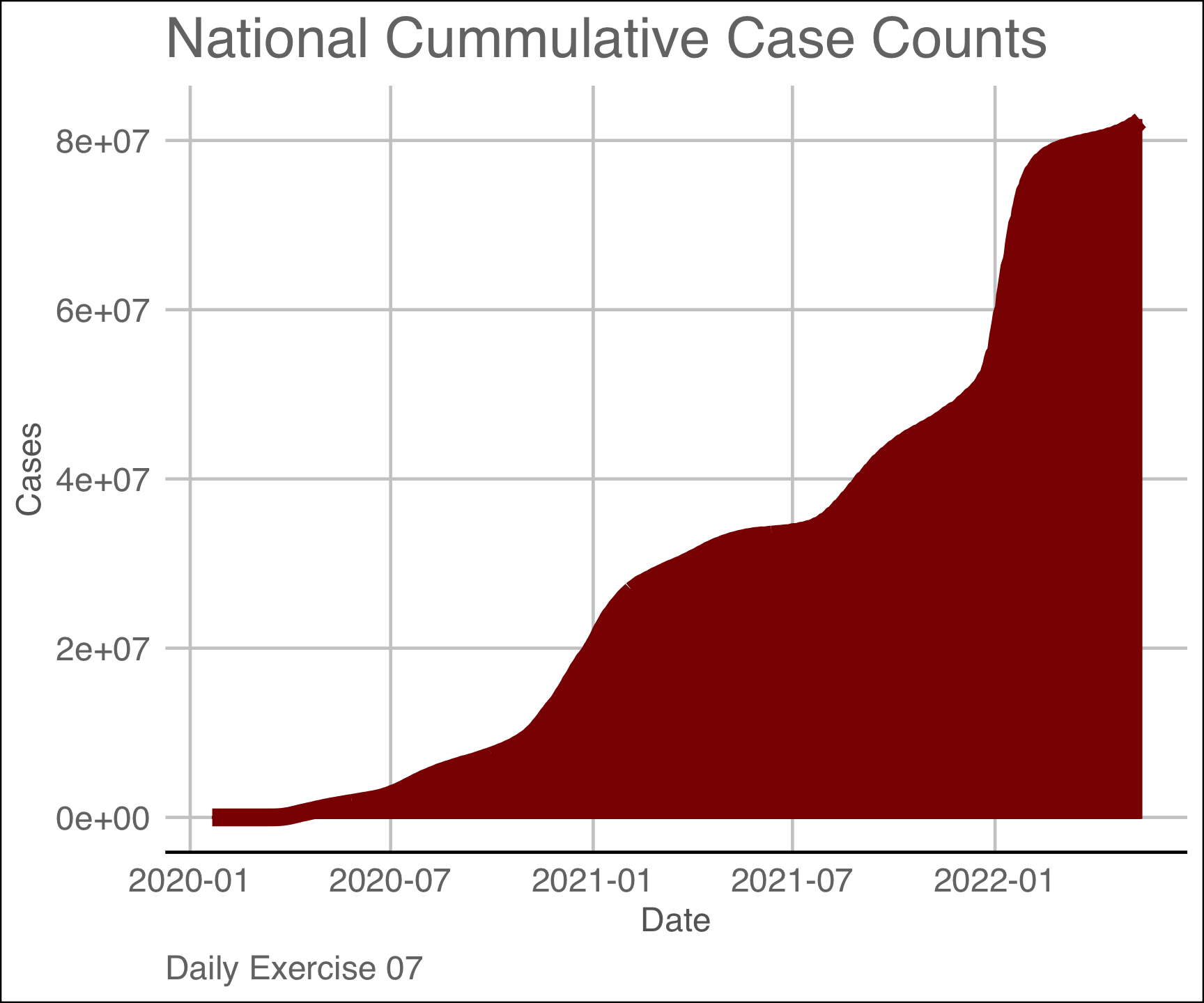
Relational Data
There will come a time when you need data from different sources.
When this happens we must join – or merge – multiple tables
Multiple tables of data are called *relational data** because the relations are an equal part of the data
To merge data, we have to find a point of commonality
That point – or attribute – of commonality is the relation
Relational Verbs
To work with relational data we need “verbs” that work with pairs (2) of tables.
Mutating joins: add new variables to one table from matching observations in another.
Filtering joins: filter observations from one table if they match an observation in the other table.
Set operations: treat observations as if they were set elements.
The most common place to find relational data is in a relational database management system (or RDBMS)
Keys
The variables used to connect a pair tables are called keys.
A key is a variable (or set of variables) that uniquely identifies an observation or “unit”.
Sometimes, a single variable is enough…
- For example, each county in the USA is uniquely identified by its FIP code.
- Each state is unique identified by its name
- …
In other cases, multiple variables may be needed.
Keys
There are two types of keys:
Primary keys: uniquely identify observations in its own table.
Foreign keys: uniquely identify observations in another table.
A primary key and a corresponding foreign key form a relation.
Relations are typically one-to-many but can be one-to-one
A quick note on surrogate keys
Sometimes a table doesn’t have an explicit primary key.
If a table lacks a primary key, it’s sometimes useful to add one with
mutate()androw_number().Doing this creates a surrogate key.
gapminder |>
slice(1:5) |>
mutate(surrogate = row_number())
# A tibble: 5 × 7
country continent year lifeExp pop gdpPercap surrogate
<fct> <fct> <int> <dbl> <int> <dbl> <int>
1 Afghanistan Asia 1952 28.8 8425333 779. 1
2 Afghanistan Asia 1957 30.3 9240934 821. 2
3 Afghanistan Asia 1962 32.0 10267083 853. 3
4 Afghanistan Asia 1967 34.0 11537966 836. 4
5 Afghanistan Asia 1972 36.1 13079460 740. 5Today’s Data:

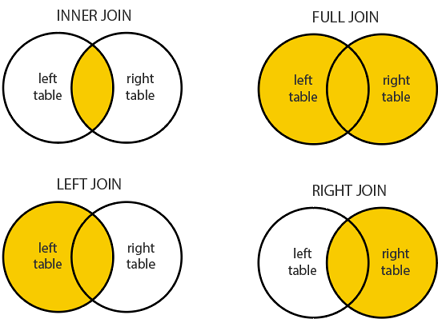
Mutating Joins
Mutating Joins add new variables to one table from matching observations in another.
1. Inner Join
inner_join(x, y): Return all rows from x where there are matching values in y, and all columns from x and y.
If there are multiple matches between x and y, all combination of the matches are returned. This is a mutating join.
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
inner_join(band_members, band_instruments, by = “name”)
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
| name | band | plays |
|---|---|---|
| John | Beatles | guitar |
| Paul | Beatles | bass |
2. Left Join
left_join(x, y): Return all rows from x, and all columns from x and y.
If there are multiple matches between x and y, all combination of the matches are returned.
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
left_join(band_members, band_instruments, by = “name”)
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
| name | band | plays |
|---|---|---|
| Mick | Stones | NA |
| John | Beatles | guitar |
| Paul | Beatles | bass |
3. Right Join
right_join(x, y): Return all rows from x where there are matching values in y, and all columns from x and y.
If there are multiple matches between x and y, all combination of the matches are returned.
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
right_join(band_members, band_instruments, by = “name”)
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
| name | band | plays |
|---|---|---|
| John | Beatles | guitar |
| Paul | Beatles | bass |
| Keith | NA | guitar |
4. Full Join
full_join(x, y): Return all rows and columns from both x and y.
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
full_join(band_members, band_instruments, by = “name”)
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
| name | band | plays |
|---|---|---|
| Mick | Stones | NA |
| John | Beatles | guitar |
| Paul | Beatles | bass |
| Keith | NA | guitar |
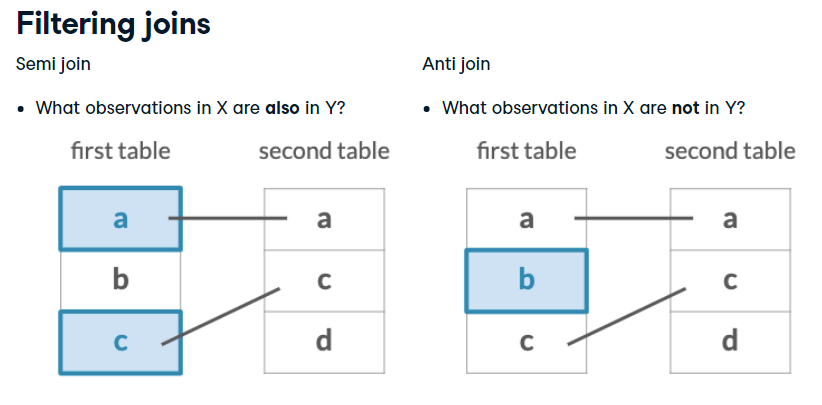
Filtering Joins
“Filtering” joins keep cases from the LHS that have a defined relation from the RHS
Keeps or removes observations from the first table
Doesn’t add new variables
Semi Join
semi_join(x, y): Return all rows from x where there are matching values in _ y_, keeping just columns from x.
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
semi_join(band_members, band_instruments, by = “name”)
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
| name | band |
|---|---|
| John | Beatles |
| Paul | Beatles |
Anti Join
anti_join(x, y): Return all rows from x where there are not matching values in y, keeping just columns from x.
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
anti_join(band_members, band_instruments, by = “name”)
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
| name | band |
|---|---|
| Mick | Stones |
When keys dont share a name
Return to the COVID example!
So far, we’ve looked at raw COVID counts. With a lot of data, raw counts are very misleading
For example does comparing the total cases in California really compare to total cases in Rhode Island given the difference in population?
Here is a perfect example of time when multiple datasets are needed to best answer a question!
Population data
The U.S. Census is the primary agency tasked with understanding our population
Google-ing for population data leads us to here
At the bottom of the page we can see their FTP (File Transfer Protocol) server
Once there, clicking on
2020-2024/state/totals/leads us to a .csv URL we can read from!
Population data
Population data
# A tibble: 66 × 75
SUMLEV REGION DIVISION STATE NAME ESTIMATESBASE2020 POPESTIMATE2020
<chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
1 010 0 0 00 United States 331515736 331577720
2 020 1 0 00 Northeast Reg… 57617706 57431458
3 030 1 1 00 New England 15122011 15057350
4 030 1 2 00 Middle Atlant… 42495695 42374108
5 020 2 0 00 Midwest Region 68998970 68984258
6 030 2 3 00 East North Ce… 47381362 47358568
7 030 2 4 00 West North Ce… 21617608 21625690
8 020 3 0 00 South Region 126281537 126476549
9 030 3 5 00 South Atlantic 66097951 66176865
10 030 3 6 00 East South Ce… 19405296 19427852
# ℹ 56 more rows
# ℹ 68 more variables: POPESTIMATE2021 <dbl>, POPESTIMATE2022 <dbl>,
# POPESTIMATE2023 <dbl>, POPESTIMATE2024 <dbl>, NPOPCHG_2020 <dbl>,
# NPOPCHG_2021 <dbl>, NPOPCHG_2022 <dbl>, NPOPCHG_2023 <dbl>,
# NPOPCHG_2024 <dbl>, BIRTHS2020 <dbl>, BIRTHS2021 <dbl>, BIRTHS2022 <dbl>,
# BIRTHS2023 <dbl>, BIRTHS2024 <dbl>, DEATHS2020 <dbl>, DEATHS2021 <dbl>,
# DEATHS2022 <dbl>, DEATHS2023 <dbl>, DEATHS2024 <dbl>, …Population data
# A tibble: 66 × 2
name pop2024
<chr> <dbl>
1 United States 340110988
2 Northeast Region 57832935
3 New England 15386085
4 Middle Atlantic 42446850
5 Midwest Region 69596584
6 East North Central 47619171
7 West North Central 21977413
8 South Region 132665693
9 South Atlantic 69676509
10 East South Central 19916866
# ℹ 56 more rowsPopulation data
# A tibble: 66 × 3
name pop2024 pop2024_per100k
<chr> <dbl> <dbl>
1 United States 340110988 3401.
2 Northeast Region 57832935 578.
3 New England 15386085 154.
4 Middle Atlantic 42446850 424.
5 Midwest Region 69596584 696.
6 East North Central 47619171 476.
7 West North Central 21977413 220.
8 South Region 132665693 1327.
9 South Atlantic 69676509 697.
10 East South Central 19916866 199.
# ℹ 56 more rowsPopulation data
Re-do the Analysis
# A tibble: 2,502,832 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2020-01-21 Snohomish Washington 53061 1 0
2 2020-01-22 Snohomish Washington 53061 1 0
3 2020-01-23 Snohomish Washington 53061 1 0
4 2020-01-24 Cook Illinois 17031 1 0
5 2020-01-24 Snohomish Washington 53061 1 0
6 2020-01-25 Orange California 06059 1 0
7 2020-01-25 Cook Illinois 17031 1 0
8 2020-01-25 Snohomish Washington 53061 1 0
9 2020-01-26 Maricopa Arizona 04013 1 0
10 2020-01-26 Los Angeles California 06037 1 0
# ℹ 2,502,822 more rowsRe-do the Analysis
# A tibble: 3,258 × 6
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2022-05-13 Autauga Alabama 01001 15863 216
2 2022-05-13 Baldwin Alabama 01003 55862 681
3 2022-05-13 Barbour Alabama 01005 5681 98
4 2022-05-13 Bibb Alabama 01007 6457 105
5 2022-05-13 Blount Alabama 01009 15005 243
6 2022-05-13 Bullock Alabama 01011 2319 54
7 2022-05-13 Butler Alabama 01013 5068 129
8 2022-05-13 Calhoun Alabama 01015 32453 627
9 2022-05-13 Chambers Alabama 01017 8508 162
10 2022-05-13 Cherokee Alabama 01019 5131 86
# ℹ 3,248 more rowsRe-do the Analysis
# A tibble: 3,258 × 6
# Groups: state [56]
date county state fips cases deaths
<date> <chr> <chr> <chr> <dbl> <dbl>
1 2022-05-13 Autauga Alabama 01001 15863 216
2 2022-05-13 Baldwin Alabama 01003 55862 681
3 2022-05-13 Barbour Alabama 01005 5681 98
4 2022-05-13 Bibb Alabama 01007 6457 105
5 2022-05-13 Blount Alabama 01009 15005 243
6 2022-05-13 Bullock Alabama 01011 2319 54
7 2022-05-13 Butler Alabama 01013 5068 129
8 2022-05-13 Calhoun Alabama 01015 32453 627
9 2022-05-13 Chambers Alabama 01017 8508 162
10 2022-05-13 Cherokee Alabama 01019 5131 86
# ℹ 3,248 more rowsRe-do the Analysis
# A tibble: 56 × 2
state cases
<chr> <dbl>
1 Alabama 1304710
2 Alaska 254467
3 American Samoa 5930
4 Arizona 2030925
5 Arkansas 838251
6 California 9351630
7 Colorado 1412121
8 Connecticut 779460
9 Delaware 267265
10 District of Columbia 143943
# ℹ 46 more rowsRe-do the Analysis
# A tibble: 56 × 2
state cases
<chr> <dbl>
1 Alabama 1304710
2 Alaska 254467
3 American Samoa 5930
4 Arizona 2030925
5 Arkansas 838251
6 California 9351630
7 Colorado 1412121
8 Connecticut 779460
9 Delaware 267265
10 District of Columbia 143943
# ℹ 46 more rowsRe-do the Analysis
# A tibble: 52 × 4
state cases pop2024 pop2024_per100k
<chr> <dbl> <dbl> <dbl>
1 Alabama 1304710 5157699 51.6
2 Alaska 254467 740133 7.40
3 Arizona 2030925 7582384 75.8
4 Arkansas 838251 3088354 30.9
5 California 9351630 39431263 394.
6 Colorado 1412121 5957493 59.6
7 Connecticut 779460 3675069 36.8
8 Delaware 267265 1051917 10.5
9 District of Columbia 143943 702250 7.02
10 Florida 5997998 23372215 234.
# ℹ 42 more rowsRe-do the Analysis
# A tibble: 52 × 5
state cases pop2024 pop2024_per100k perCapCases
<chr> <dbl> <dbl> <dbl> <dbl>
1 Alabama 1304710 5157699 51.6 25296.
2 Alaska 254467 740133 7.40 34381.
3 Arizona 2030925 7582384 75.8 26785.
4 Arkansas 838251 3088354 30.9 27142.
5 California 9351630 39431263 394. 23716.
6 Colorado 1412121 5957493 59.6 23703.
7 Connecticut 779460 3675069 36.8 21209.
8 Delaware 267265 1051917 10.5 25407.
9 District of Columbia 143943 702250 7.02 20497.
10 Florida 5997998 23372215 234. 25663.
# ℹ 42 more rowsRe-do the Analysis
# A tibble: 6 × 5
state cases pop2024 pop2024_per100k perCapCases
<chr> <dbl> <dbl> <dbl> <dbl>
1 Alaska 254467 740133 7.40 34381.
2 Rhode Island 381271 1112308 11.1 34277.
3 North Dakota 242462 796568 7.97 30438.
4 Kentucky 1345754 4588372 45.9 29330.
5 West Virginia 505528 1769979 17.7 28561.
6 Tennessee 1990937 7227750 72.3 27546.Re-do the Analysis
Re-do the Analysis
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
inner_join(pop, by = c("state" = "name")) |>
mutate(perCapCases = cases / pop2024_per100k) |>
slice_max(perCapCases, n = 6) |>
ggplot() +
geom_col(aes(x = reorder(state, -perCapCases), y = perCapCases))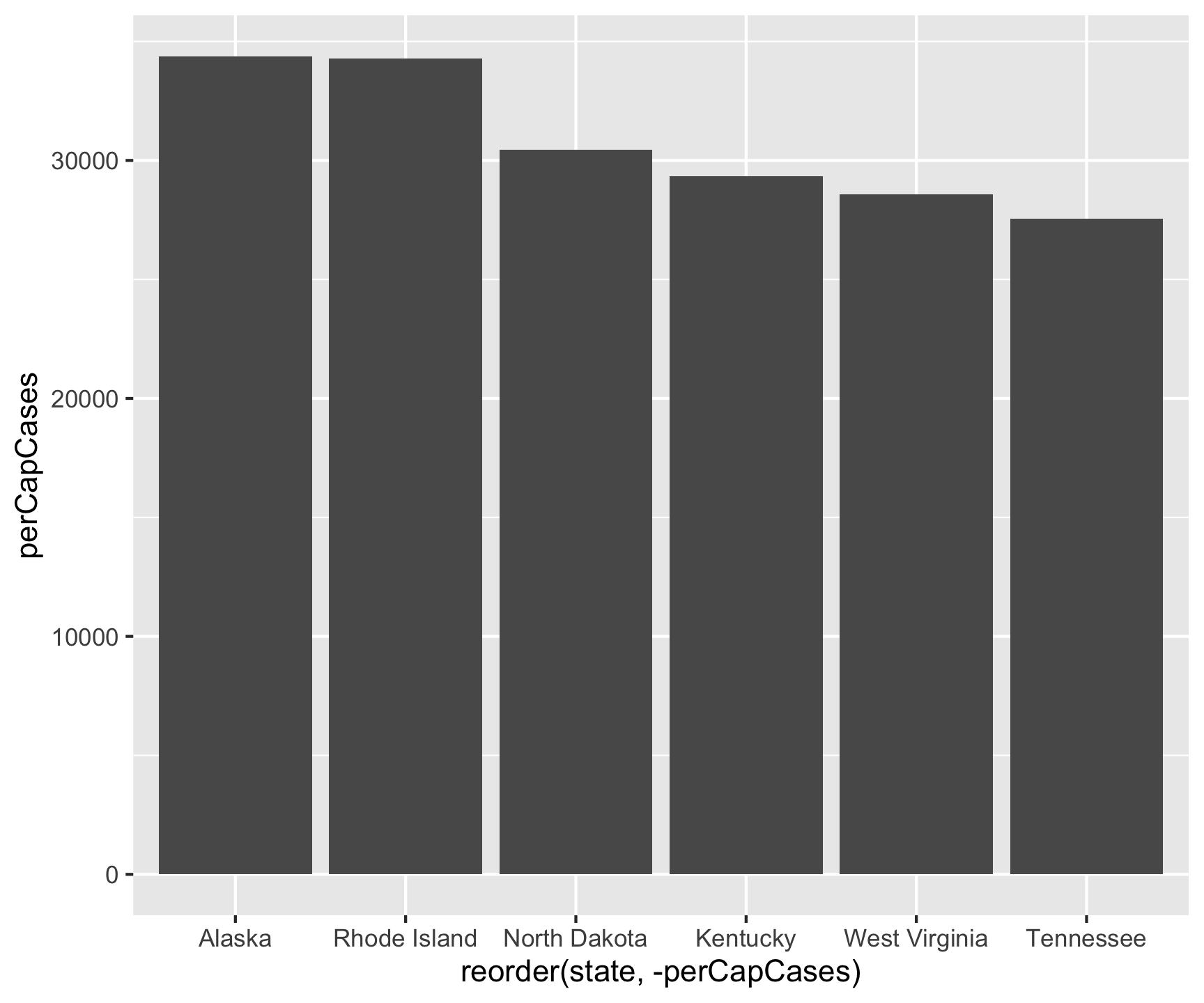
Re-do the Analysis
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
inner_join(pop, by = c("state" = "name")) |>
mutate(perCapCases = cases / pop2024_per100k) |>
slice_max(perCapCases, n = 6) |>
ggplot() +
geom_col(aes(x = reorder(state, -perCapCases), y = perCapCases)) +
labs(title = "Covid Cases per 100k (Top States)",
x = "State",
y = "Cases per 100k population")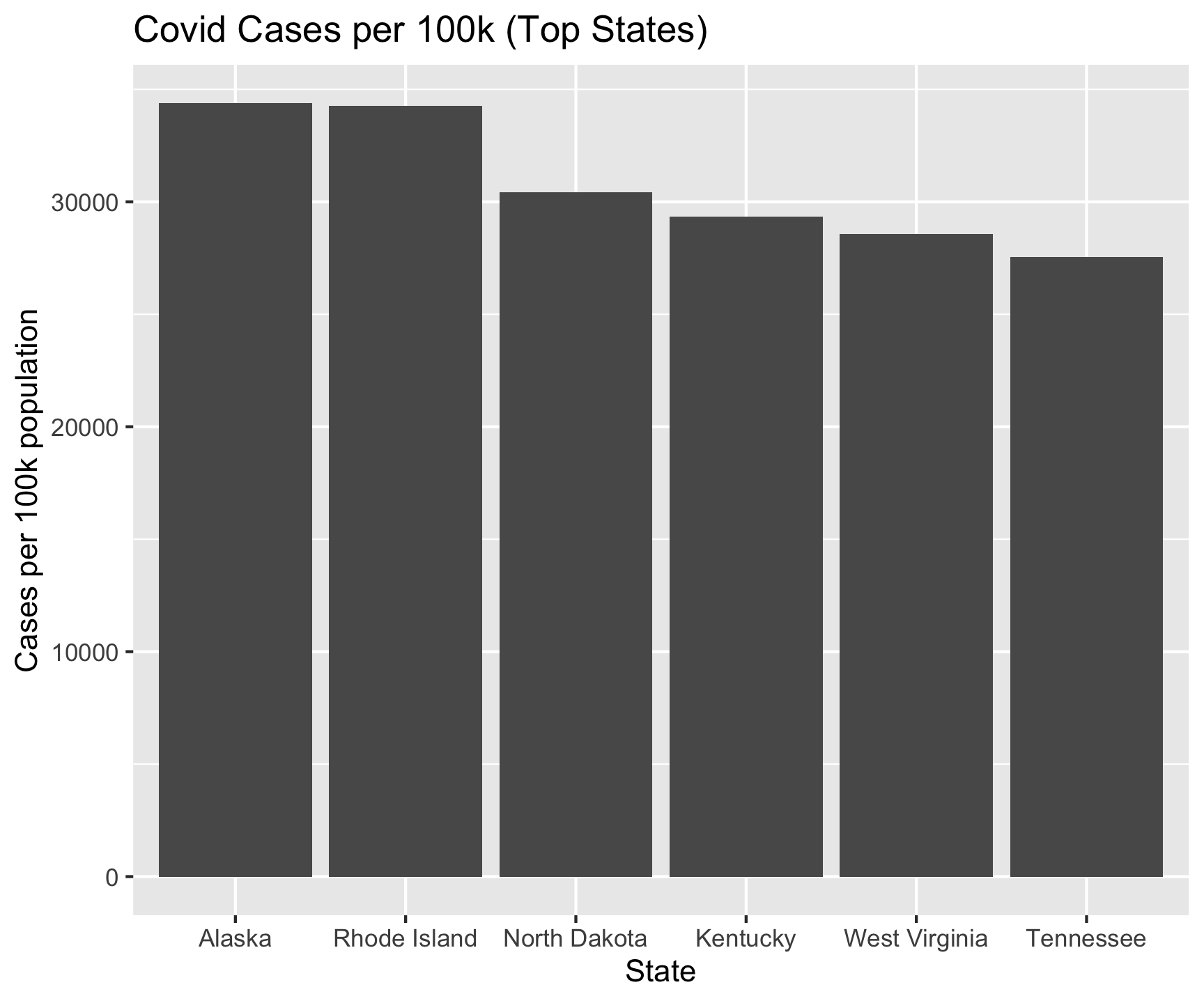
Re-do the Analysis
covid |>
filter(date == max(date)) |>
group_by(state) |>
summarize(cases = sum(cases, na.rm = TRUE)) |>
ungroup() |>
inner_join(pop, by = c("state" = "name")) |>
mutate(perCapCases = cases / pop2024_per100k) |>
slice_max(perCapCases, n = 6) |>
ggplot() +
geom_col(aes(x = reorder(state, -perCapCases), y = perCapCases)) +
labs(title = "Covid Cases per 100k (Top States)",
x = "State",
y = "Cases per 100k population") +
theme_linedraw()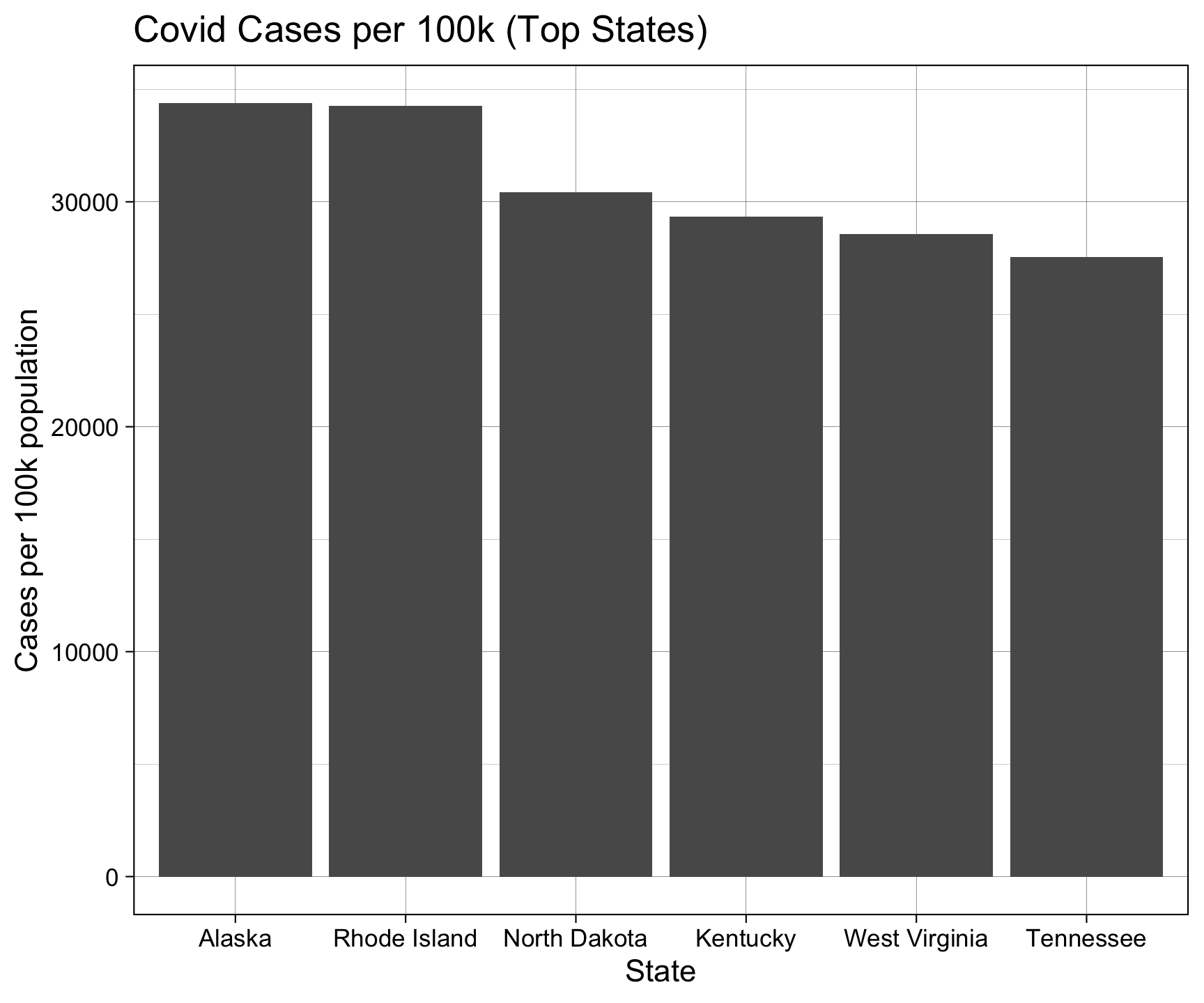
Data structure vs Data format
data.framesare a list with atomic vectors of the same length.data.frames(and list and vector) are data structuresThe format of the
data.framehowever can be widely variable….
For example …
You can represent the same underlying data in multiple ways. Each of these stores data for country, population and cases in a different way….
country | year | type | value |
|---|---|---|---|
Brazil | 1,952 | lifeExp | 50.9170 |
Brazil | 1,952 | gdpPercap | 2,108.9444 |
Brazil | 2,007 | lifeExp | 72.3900 |
Brazil | 2,007 | gdpPercap | 9,065.8008 |
India | 1,952 | lifeExp | 37.3730 |
India | 1,952 | gdpPercap | 546.5657 |
India | 2,007 | lifeExp | 64.6980 |
India | 2,007 | gdpPercap | 2,452.2104 |
country | year | values |
|---|---|---|
Brazil | 1,952 | 50.917 / 2108.944355 |
Brazil | 2,007 | 72.39 / 9065.800825 |
India | 1,952 | 37.373 / 546.5657493 |
India | 2,007 | 64.698 / 2452.210407 |
country | year | lifeExp | gdpPercap |
|---|---|---|---|
Brazil | 1,952 | 50.917 | 2,108.9444 |
Brazil | 2,007 | 72.390 | 9,065.8008 |
India | 1,952 | 37.373 | 546.5657 |
India | 2,007 | 64.698 | 2,452.2104 |
country | 1952 | 2007 |
|---|---|---|
Brazil | 2,108.9444 | 9,065.801 |
India | 546.5657 | 2,452.210 |
country | 1952 | 2007 |
|---|---|---|
Brazil | 50.917 | 72.390 |
India | 37.373 | 64.698 |
Data Format – the quest for “tidy data”
These are all representations of the same underlying data, but they are not equally easy to use. One dataset, the tidy dataset, will be much easier to work with inside the tidyverse.
Tidy data demands three interrelated rules
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.

It’s impossible to only satisfy \(2/3\) so … an simpler guideline:
- Put each dataset in a tibble.
- Put each variable in a column.
BUT … the world is messy …
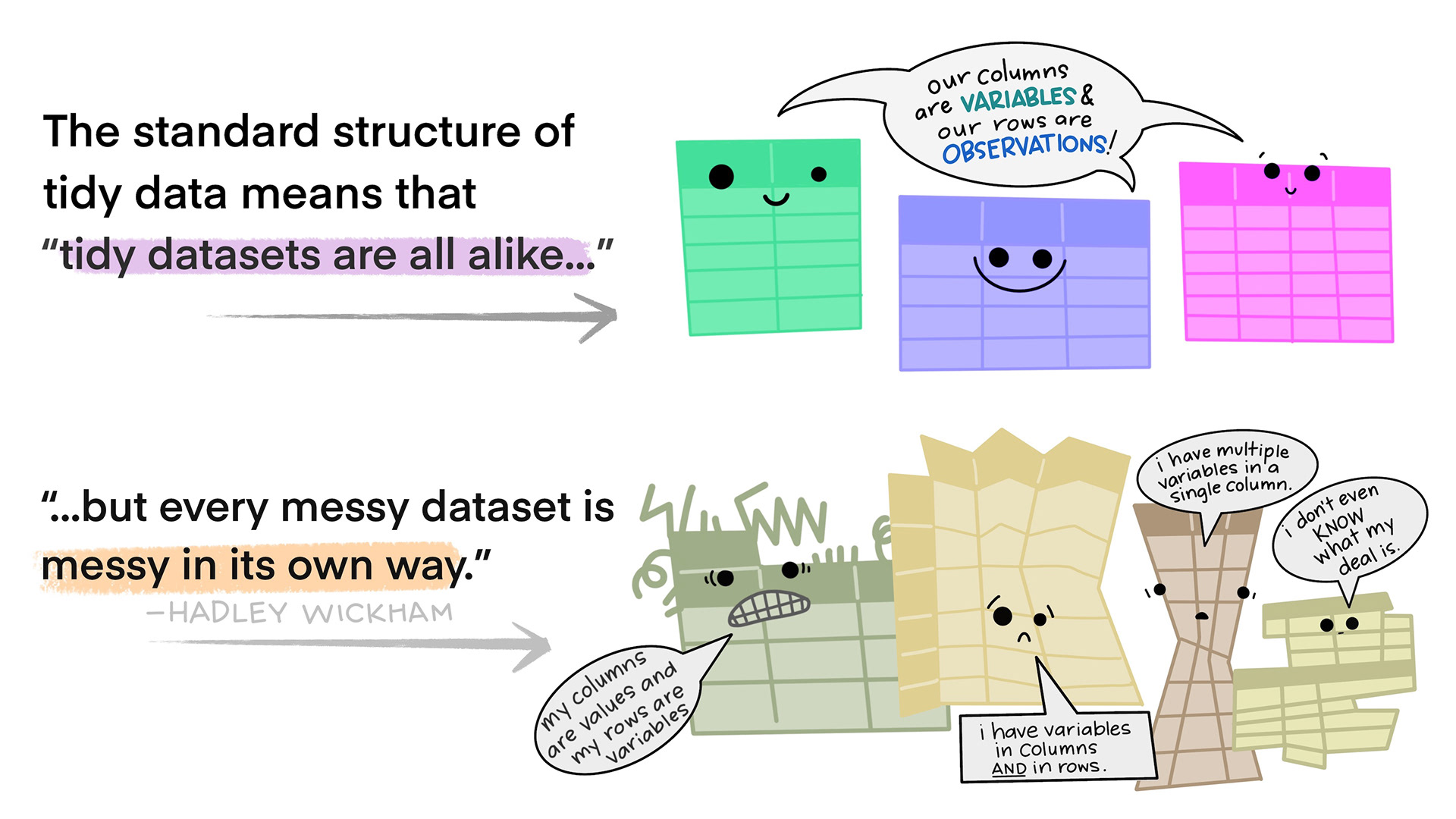
- For most real problems, you’ll need to do some cleaning/tidying …
- Once you understand what data you have, you typically have to resolve one of two common issues:
- One variable might be spread across multiple columns.
- One observation might be scattered across multiple rows.
Longer
- A common issue is a dataset where some of the columns are not names, but rather values we want:
country | 1952 | 2007 |
|---|---|---|
Brazil | 50.917 | 72.390 |
India | 37.373 | 64.698 |
Longer (and narrower)
- In long format, each observation is recorded in a single row, with a column for the variable and another for the value.
- To make the column names an variable, we need to pivot the data making the data longer and narrower…
- To do this we look to
tidyr::pivot_longer()
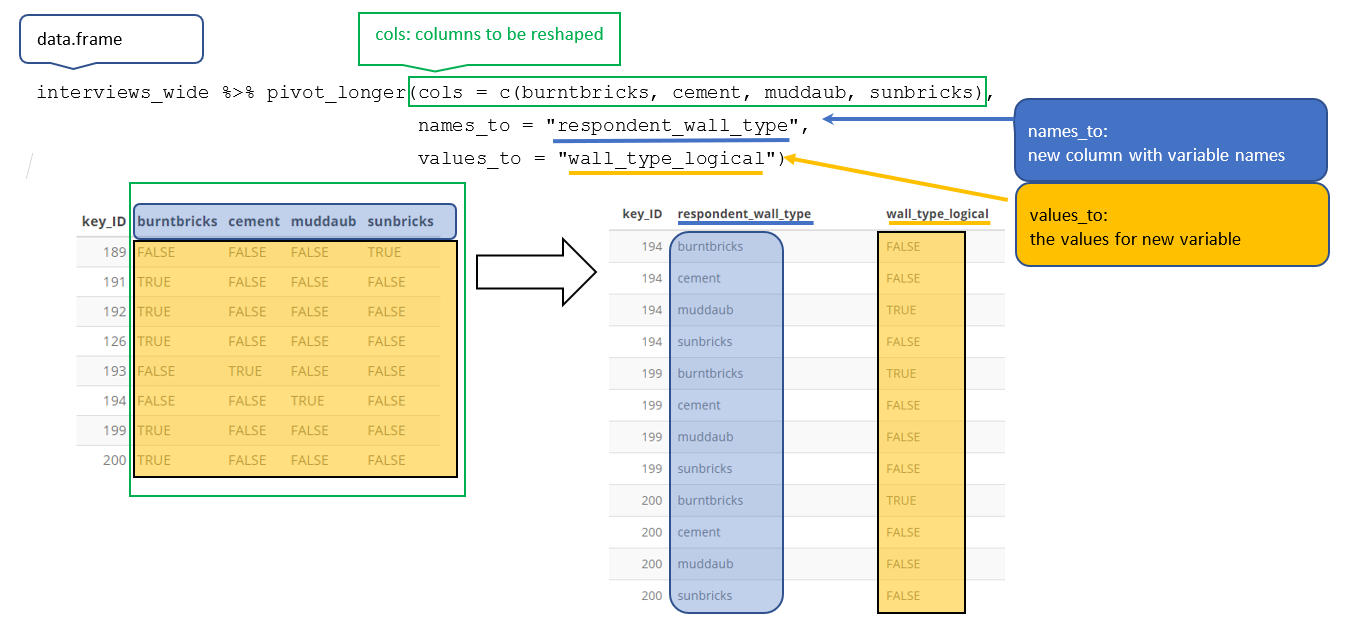
Longer
Wider (and shorter)
pivot_wider()is the opposite ofpivot_longer().We use it to solve the opposite problem… when an observation is scattered across multiple rows.
Wider (and shorter)
- To allow an variable to become a set of columns, we need to pivot the data making the data wider and shorter
- To do this we use
tidyr::pivot_wider()
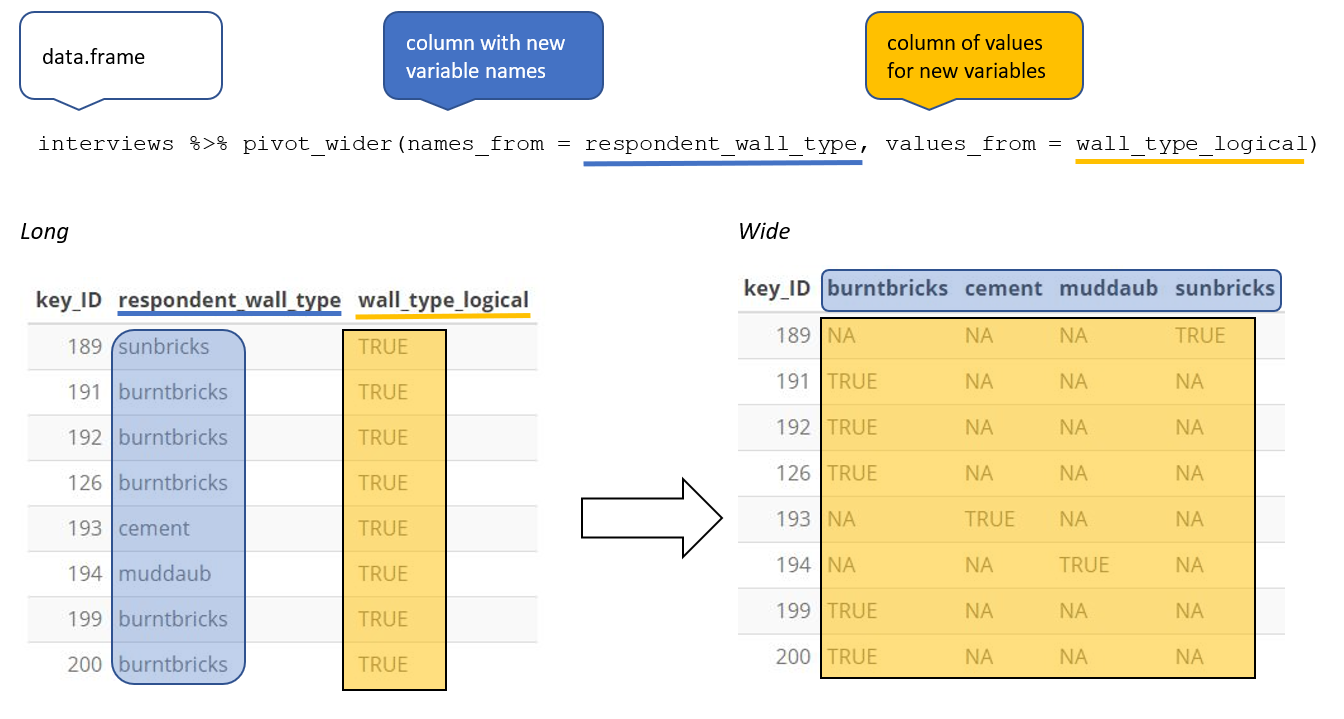
Wider
Example
- Remember the idea of “binding”? Where a set of values are “bound” to a name?
You job: - Always consider what - for your analysis - is a name and what is data
Often your needs don’t match the way data was collected/curated by others!
Your needs can range dramatically within a workflow as well
- What needs to be manipulated?
- How do you establish keys?
- What is needed for visualization?
- …
Example
Wide
# All country/Year/GDP combinations are unique rows
table_wide
# A tibble: 4 × 4
country year lifeExp gdpPercap
<fct> <int> <dbl> <dbl>
1 Brazil 1952 50.9 2109.
2 Brazil 2007 72.4 9066.
3 India 1952 37.4 547.
4 India 2007 64.7 2452.
table_wide |>
ggplot(aes(x = year, y = gdpPercap)) +
geom_line(aes(color = country)) +
facet_wrap(~country) +
theme_bw() +
theme(legend.position = "none")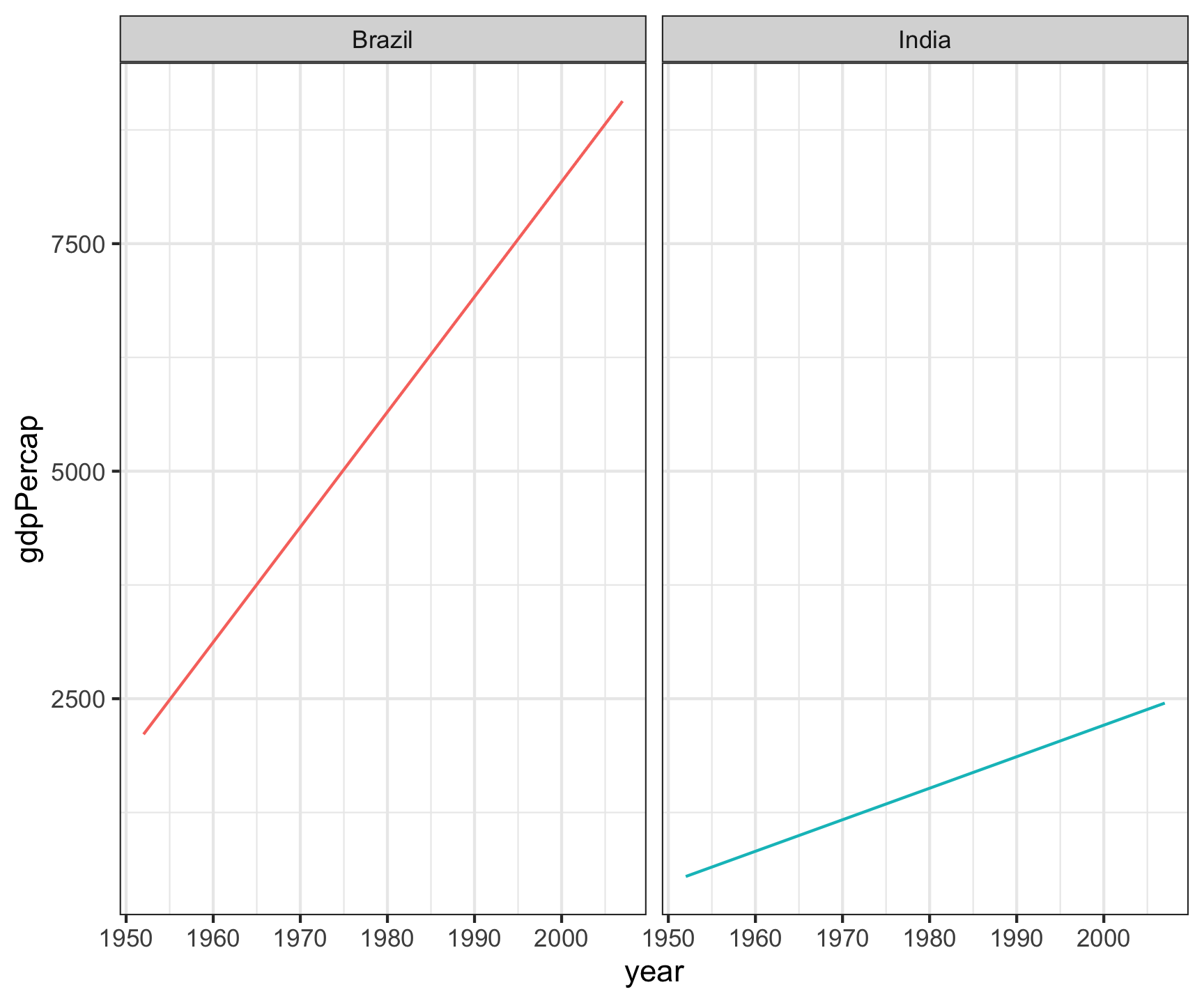
Long
# All country/Year/LifeExp/GDP combinations are unique rows
table_long
# A tibble: 8 × 4
country year type value
<fct> <int> <chr> <dbl>
1 Brazil 1952 lifeExp 50.9
2 Brazil 1952 gdpPercap 2109.
3 Brazil 2007 lifeExp 72.4
4 Brazil 2007 gdpPercap 9066.
5 India 1952 lifeExp 37.4
6 India 1952 gdpPercap 547.
7 India 2007 lifeExp 64.7
8 India 2007 gdpPercap 2452.
table_long |>
ggplot(aes(x = year, y = value)) +
geom_line(aes(color = country)) +
facet_grid(type~country, scales = "free_y") +
theme_bw() +
theme(legend.position = "none")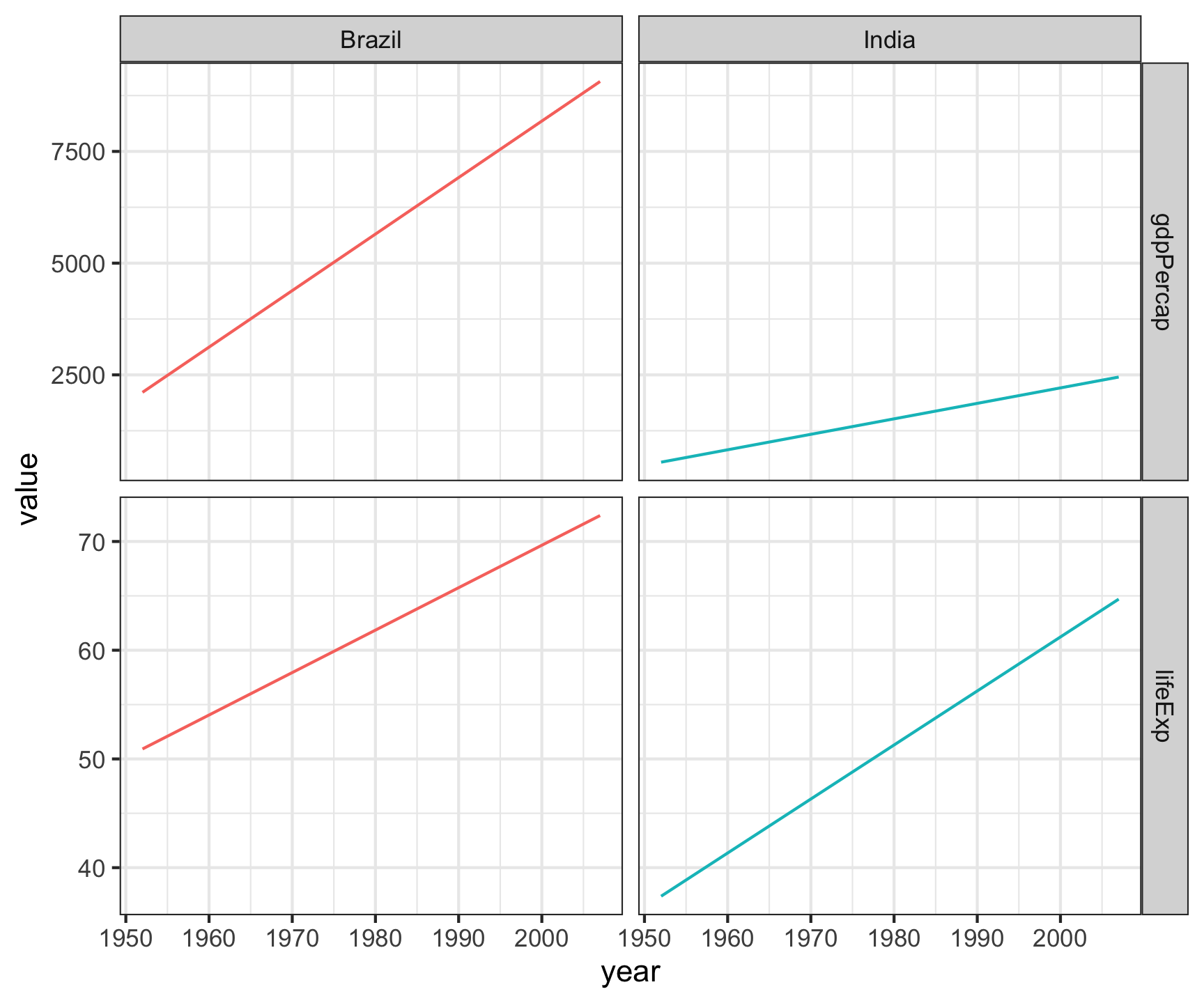
Pivot within workflow…
# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsPivot within workflow…
# A tibble: 24 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Canada Americas 1952 68.8 14785584 11367.
2 Canada Americas 1957 70.0 17010154 12490.
3 Canada Americas 1962 71.3 18985849 13462.
4 Canada Americas 1967 72.1 20819767 16077.
5 Canada Americas 1972 72.9 22284500 18971.
6 Canada Americas 1977 74.2 23796400 22091.
7 Canada Americas 1982 75.8 25201900 22899.
8 Canada Americas 1987 76.9 26549700 26627.
9 Canada Americas 1992 78.0 28523502 26343.
10 Canada Americas 1997 78.6 30305843 28955.
# ℹ 14 more rowsPivot within workflow…
# A tibble: 24 × 4
country year lifeExp gdpPercap
<fct> <int> <dbl> <dbl>
1 Canada 1952 68.8 11367.
2 Canada 1957 70.0 12490.
3 Canada 1962 71.3 13462.
4 Canada 1967 72.1 16077.
5 Canada 1972 72.9 18971.
6 Canada 1977 74.2 22091.
7 Canada 1982 75.8 22899.
8 Canada 1987 76.9 26627.
9 Canada 1992 78.0 26343.
10 Canada 1997 78.6 28955.
# ℹ 14 more rowsPivot within workflow…
# A tibble: 48 × 4
country year name value
<fct> <int> <chr> <dbl>
1 Canada 1952 lifeExp 68.8
2 Canada 1952 gdpPercap 11367.
3 Canada 1957 lifeExp 70.0
4 Canada 1957 gdpPercap 12490.
5 Canada 1962 lifeExp 71.3
6 Canada 1962 gdpPercap 13462.
7 Canada 1967 lifeExp 72.1
8 Canada 1967 gdpPercap 16077.
9 Canada 1972 lifeExp 72.9
10 Canada 1972 gdpPercap 18971.
# ℹ 38 more rowsPivot within workflow…
Pivot within workflow…
Pivot within workflow…
Pivot within workflow…
gapminder |>
filter(country %in%
c("Canada", "United States")) |>
select(country, year, lifeExp, gdpPercap) |>
pivot_longer(cols = c('lifeExp', 'gdpPercap')) |>
ggplot(aes(x = year, y = value)) +
geom_line(col = 'gray80') +
geom_point(aes(col = country)) +
facet_grid(name~country, scales = "free_y")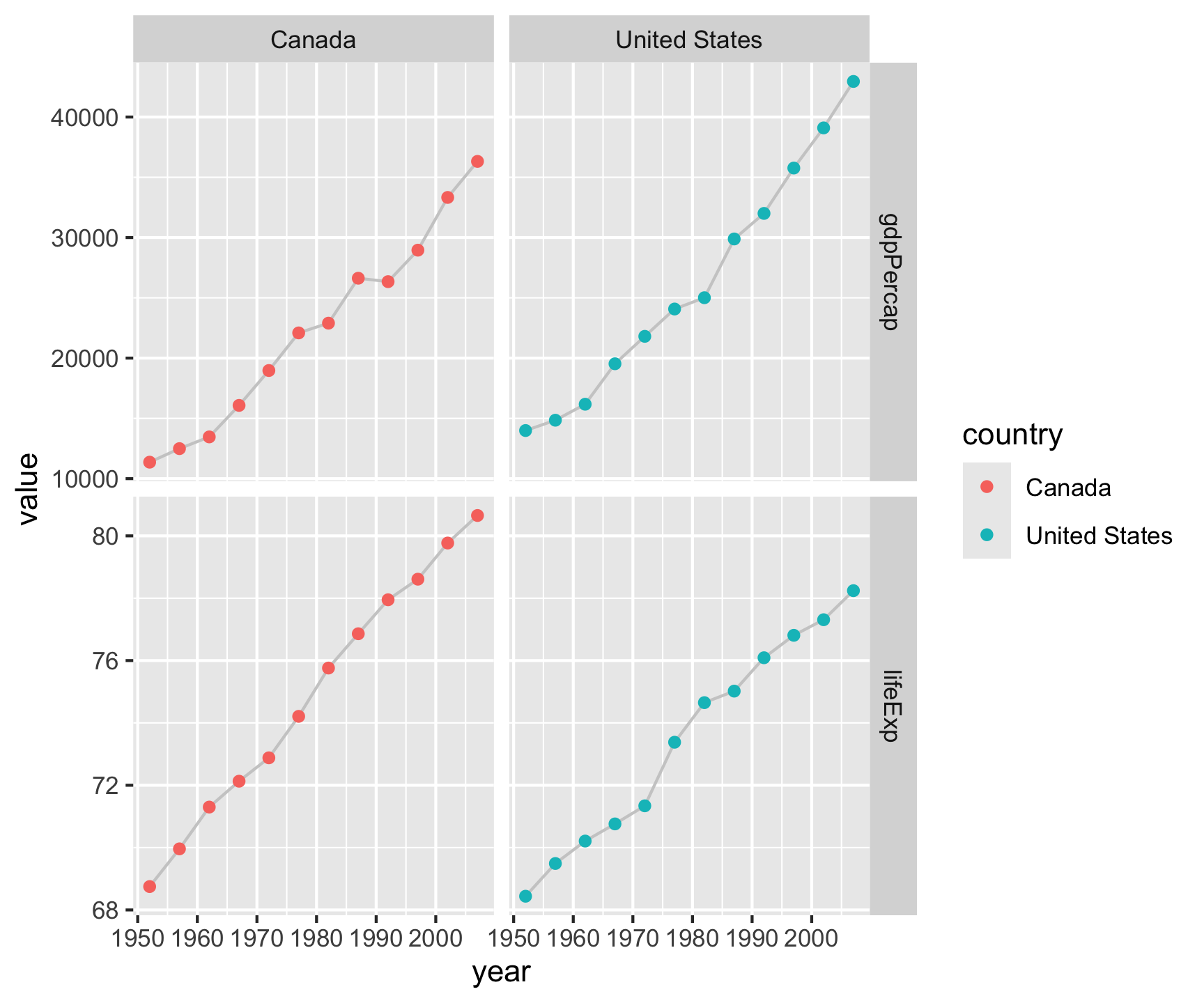
Pivot within workflow…
gapminder |>
filter(country %in%
c("Canada", "United States")) |>
select(country, year, lifeExp, gdpPercap) |>
pivot_longer(cols = c('lifeExp', 'gdpPercap')) |>
ggplot(aes(x = year, y = value)) +
geom_line(col = 'gray80') +
geom_point(aes(col = country)) +
facet_grid(name~country, scales = "free_y") +
labs(x = "", y = "Year",
title = "North America: GDP & Life Expectancy",
subtitle = "1950-2007",
caption = "Data: Gapminder R package")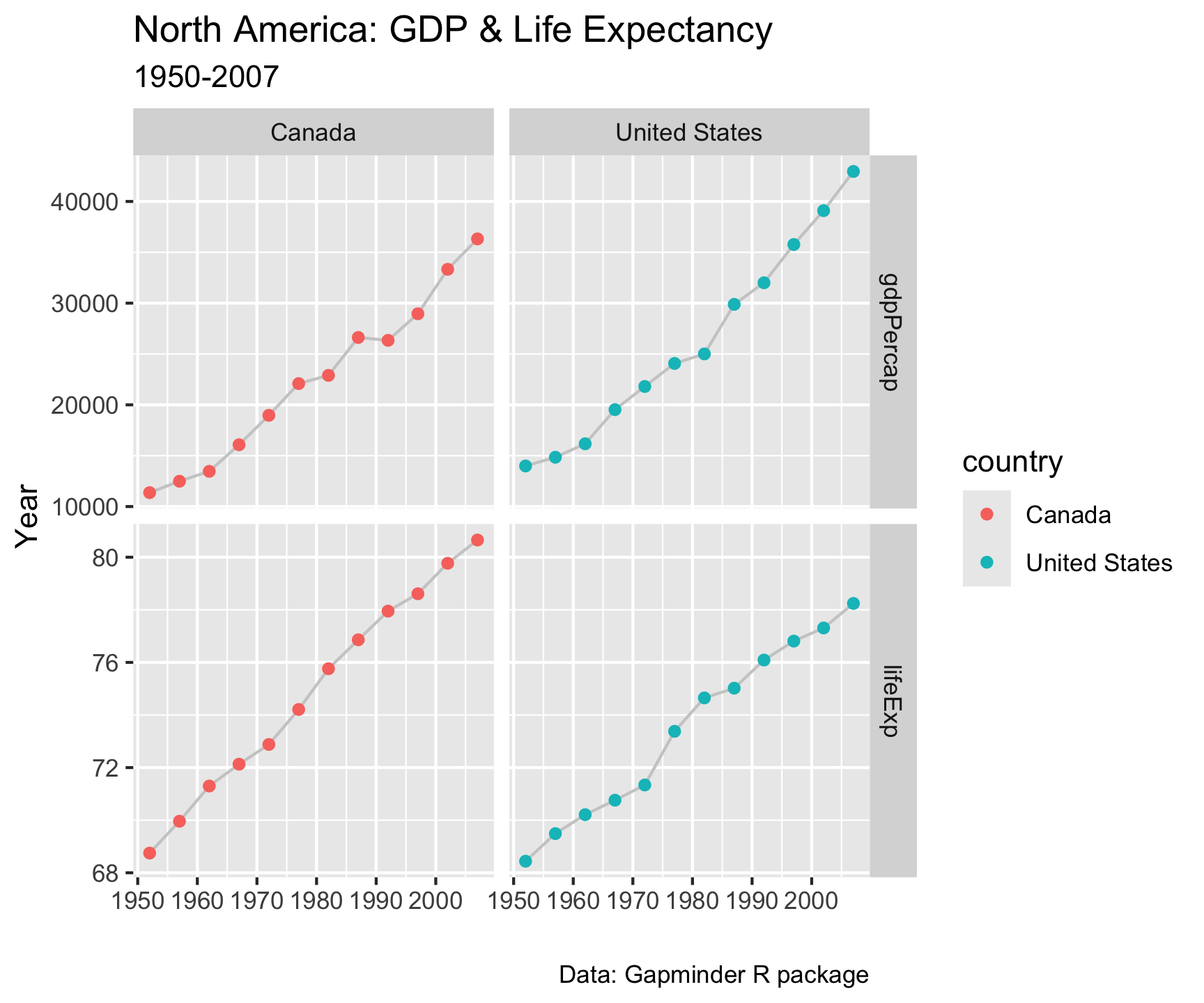
Pivot within workflow…
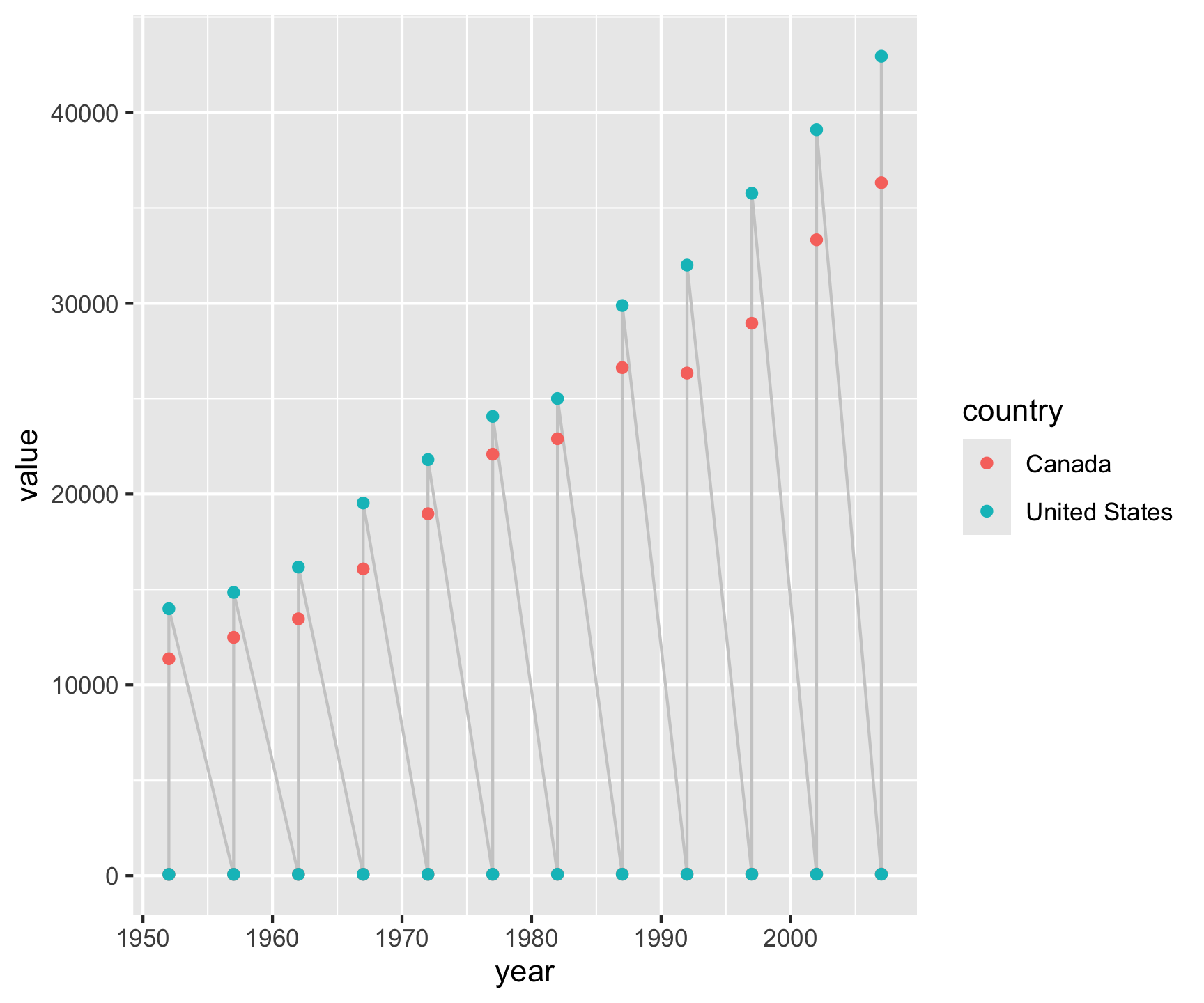
gapminder |>
filter(country %in%
c("Canada", "United States")) |>
select(country, year, lifeExp, gdpPercap) |>
pivot_longer(cols = c('lifeExp', 'gdpPercap')) |>
ggplot(aes(x = year, y = value)) +
geom_line(col = 'gray80') +
geom_point(aes(col = country)) +
facet_grid(name~country, scales = "free_y") +
labs(x = "", y = "Year",
title = "North America: GDP & Life Expectancy",
subtitle = "1950-2007",
caption = "Data: Gapminder R package") +
theme_linedraw()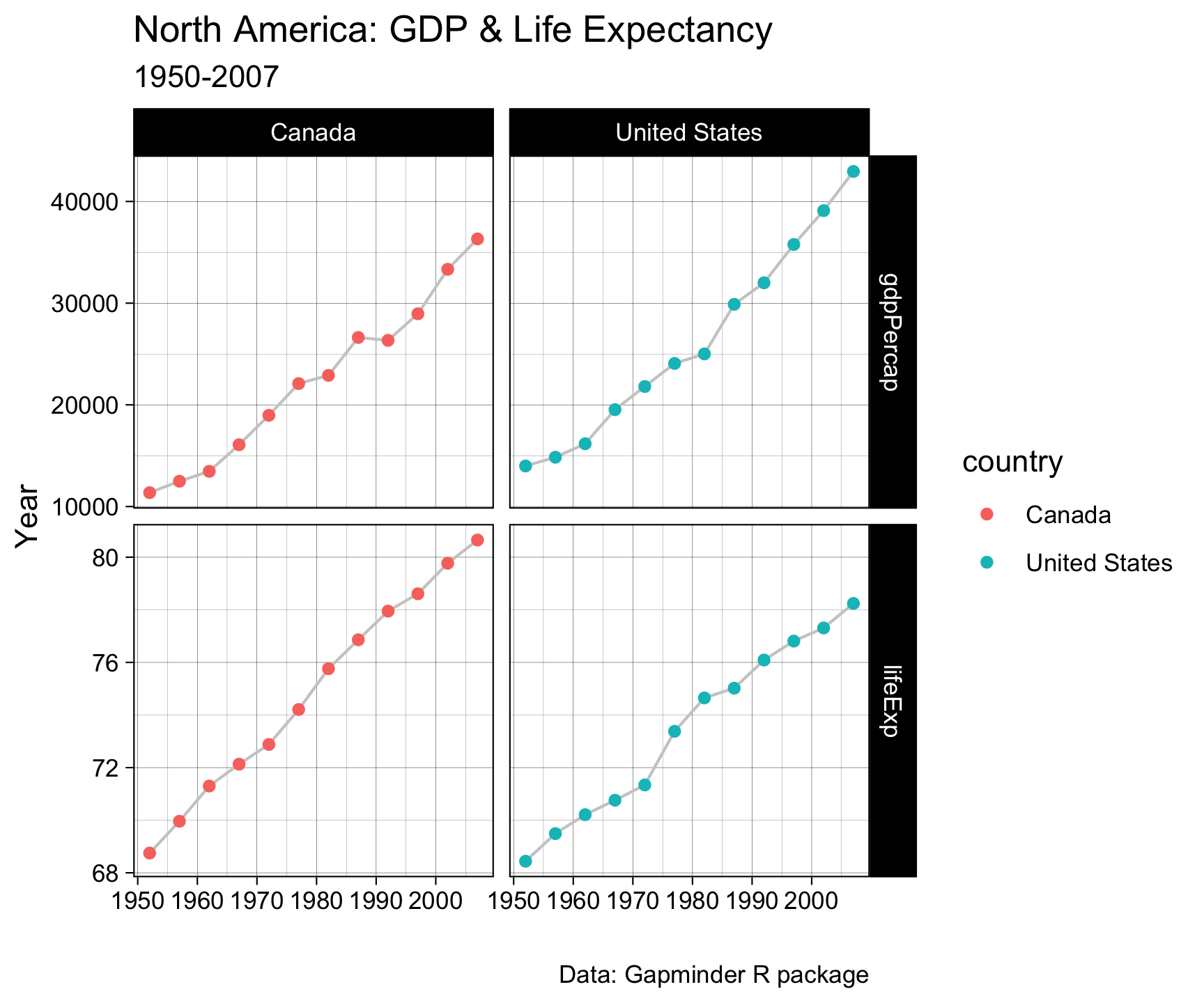
Pivot within workflow…
gapminder |>
filter(country %in%
c("Canada", "United States")) |>
select(country, year, lifeExp, gdpPercap) |>
pivot_longer(cols = c('lifeExp', 'gdpPercap')) |>
ggplot(aes(x = year, y = value)) +
geom_line(col = 'gray80') +
geom_point(aes(col = country)) +
facet_grid(name~country, scales = "free_y") +
labs(x = "", y = "Year",
title = "North America: GDP & Life Expectancy",
subtitle = "1950-2007",
caption = "Data: Gapminder R package") +
theme_linedraw() +
theme(legend.position = "none")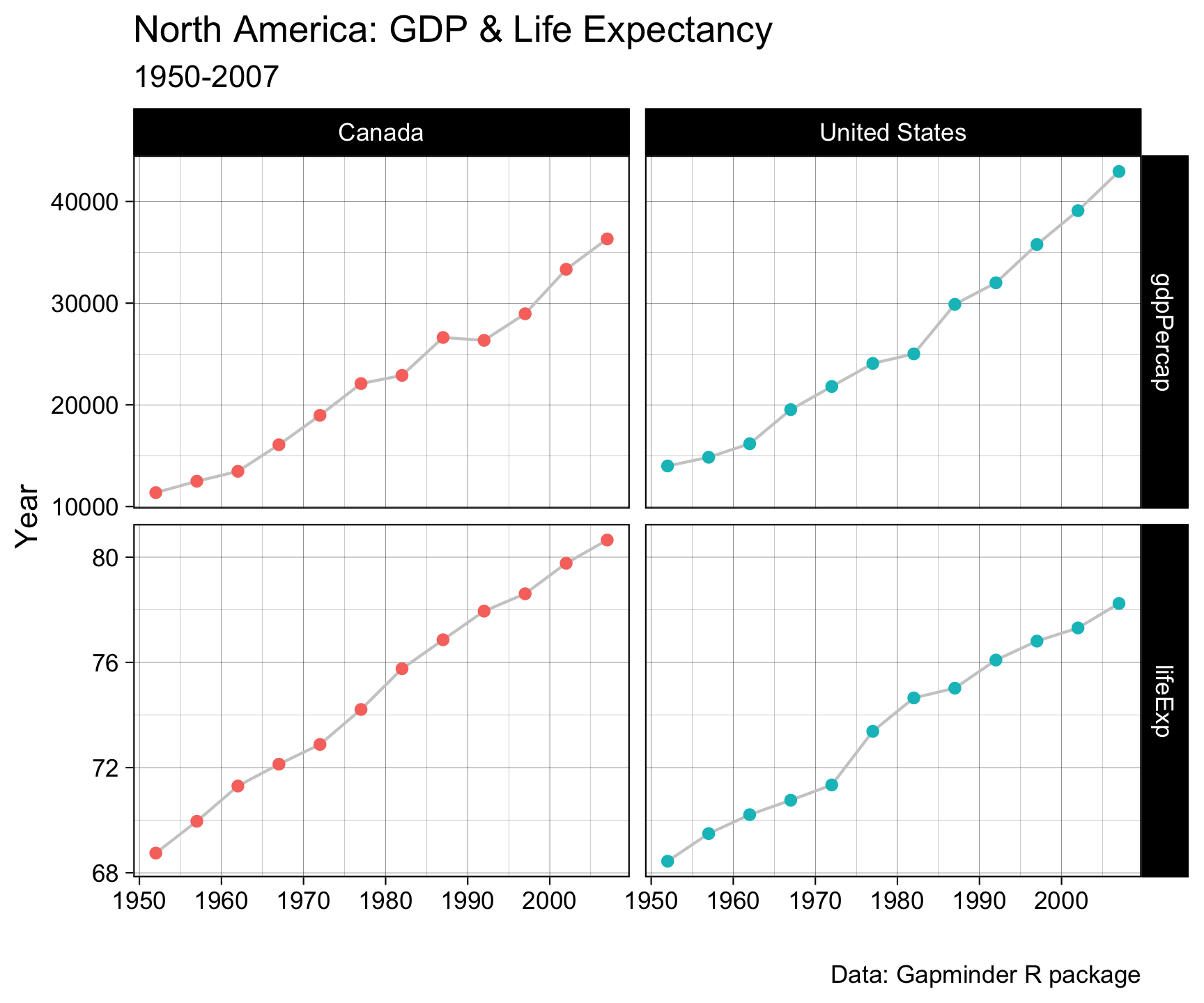
Pivot within workflow…
gapminder |>
filter(country %in%
c("Canada", "United States")) |>
select(country, year, lifeExp, gdpPercap) |>
pivot_longer(cols = c('lifeExp', 'gdpPercap')) |>
ggplot(aes(x = year, y = value)) +
geom_line(col = 'gray80') +
geom_point(aes(col = country)) +
facet_grid(name~country, scales = "free_y") +
labs(x = "", y = "Year",
title = "North America: GDP & Life Expectancy",
subtitle = "1950-2007",
caption = "Data: Gapminder R package") +
theme_linedraw() +
theme(legend.position = "none") +
theme(axis.text.x = element_text(angle = 90, face = "bold"))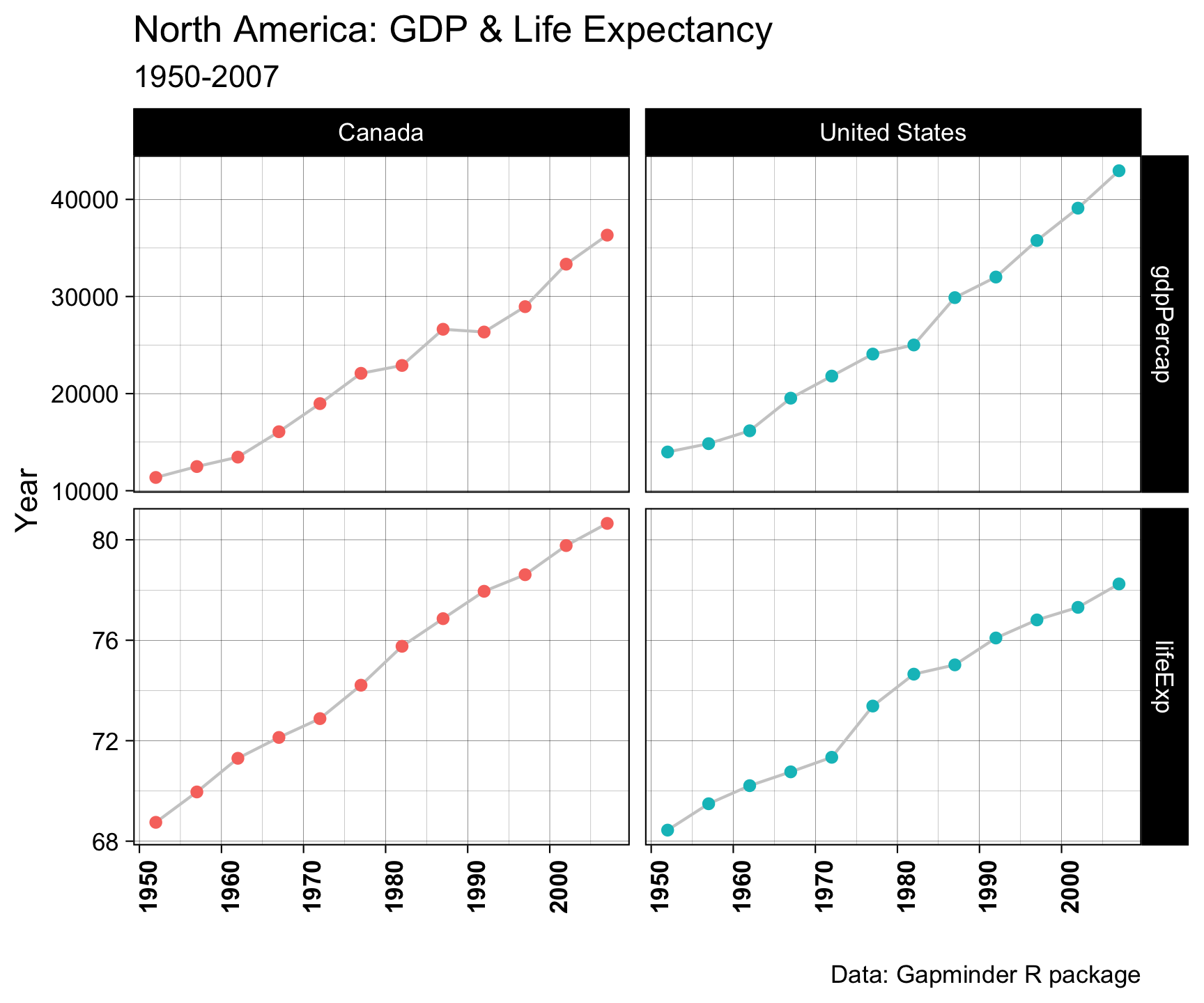
Pivot within workflow…
gapminder |>
filter(country %in%
c("Canada", "United States")) |>
select(country, year, lifeExp, gdpPercap) |>
pivot_longer(cols = c('lifeExp', 'gdpPercap')) |>
ggplot(aes(x = year, y = value)) +
geom_line(col = 'gray80') +
geom_point(aes(col = country)) +
facet_grid(name~country, scales = "free_y") +
labs(x = "", y = "Year",
title = "North America: GDP & Life Expectancy",
subtitle = "1950-2007",
caption = "Data: Gapminder R package") +
theme_linedraw() +
theme(legend.position = "none") +
theme(axis.text.x = element_text(angle = 90, face = "bold")) +
theme(plot.subtitle = element_text(color = "navy", face = "bold"))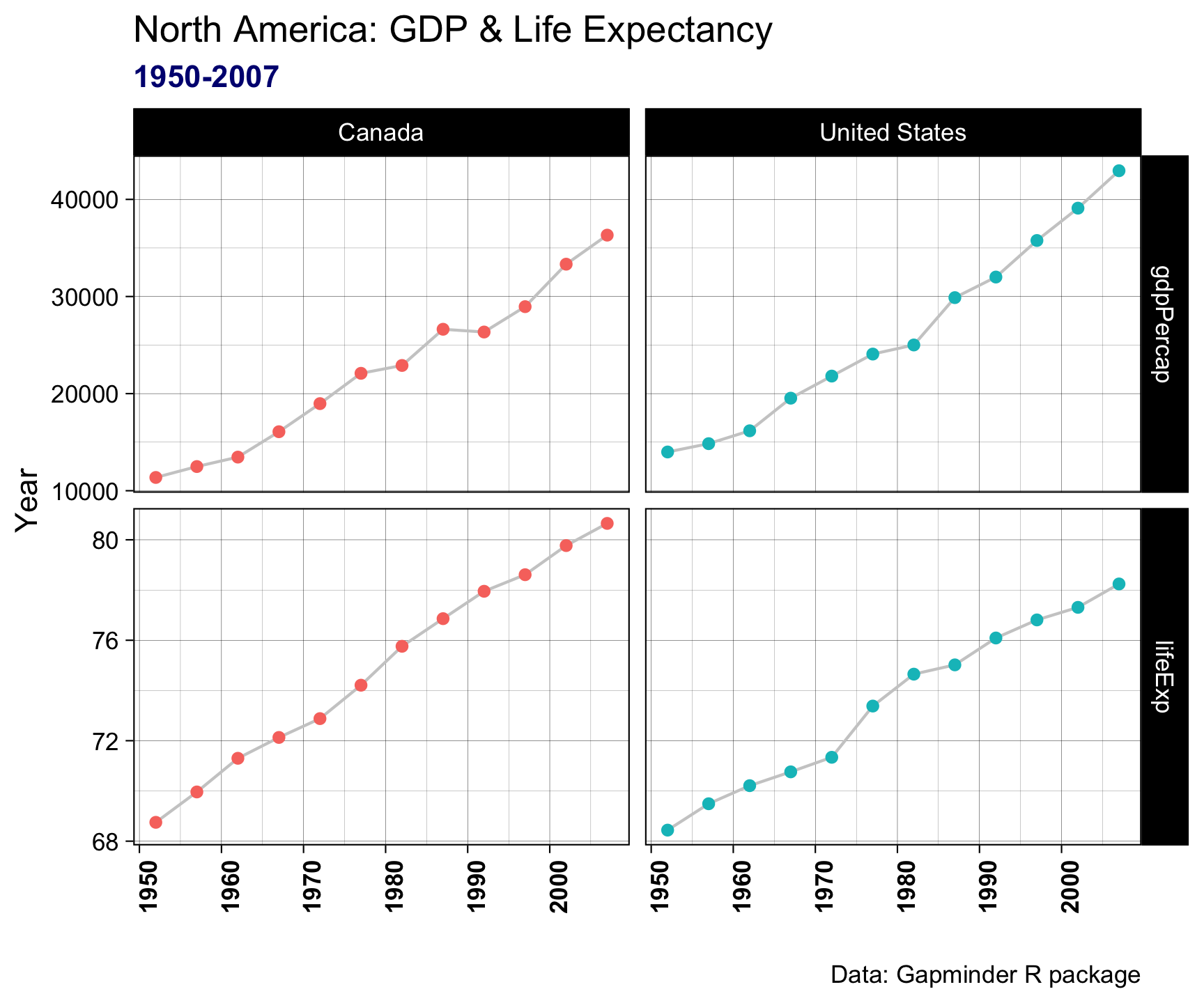
Pivot within workflow…
gapminder |>
filter(country %in%
c("Canada", "United States")) |>
select(country, year, lifeExp, gdpPercap) |>
pivot_longer(cols = c('lifeExp', 'gdpPercap')) |>
ggplot(aes(x = year, y = value)) +
geom_line(col = 'gray80') +
geom_point(aes(col = country)) +
facet_grid(name~country, scales = "free_y") +
labs(x = "", y = "Year",
title = "North America: GDP & Life Expectancy",
subtitle = "1950-2007",
caption = "Data: Gapminder R package") +
theme_linedraw() +
theme(legend.position = "none") +
theme(axis.text.x = element_text(angle = 90, face = "bold")) +
theme(plot.subtitle = element_text(color = "navy", face = "bold")) +
theme(plot.caption = element_text(color = "gray50", face = "italic"))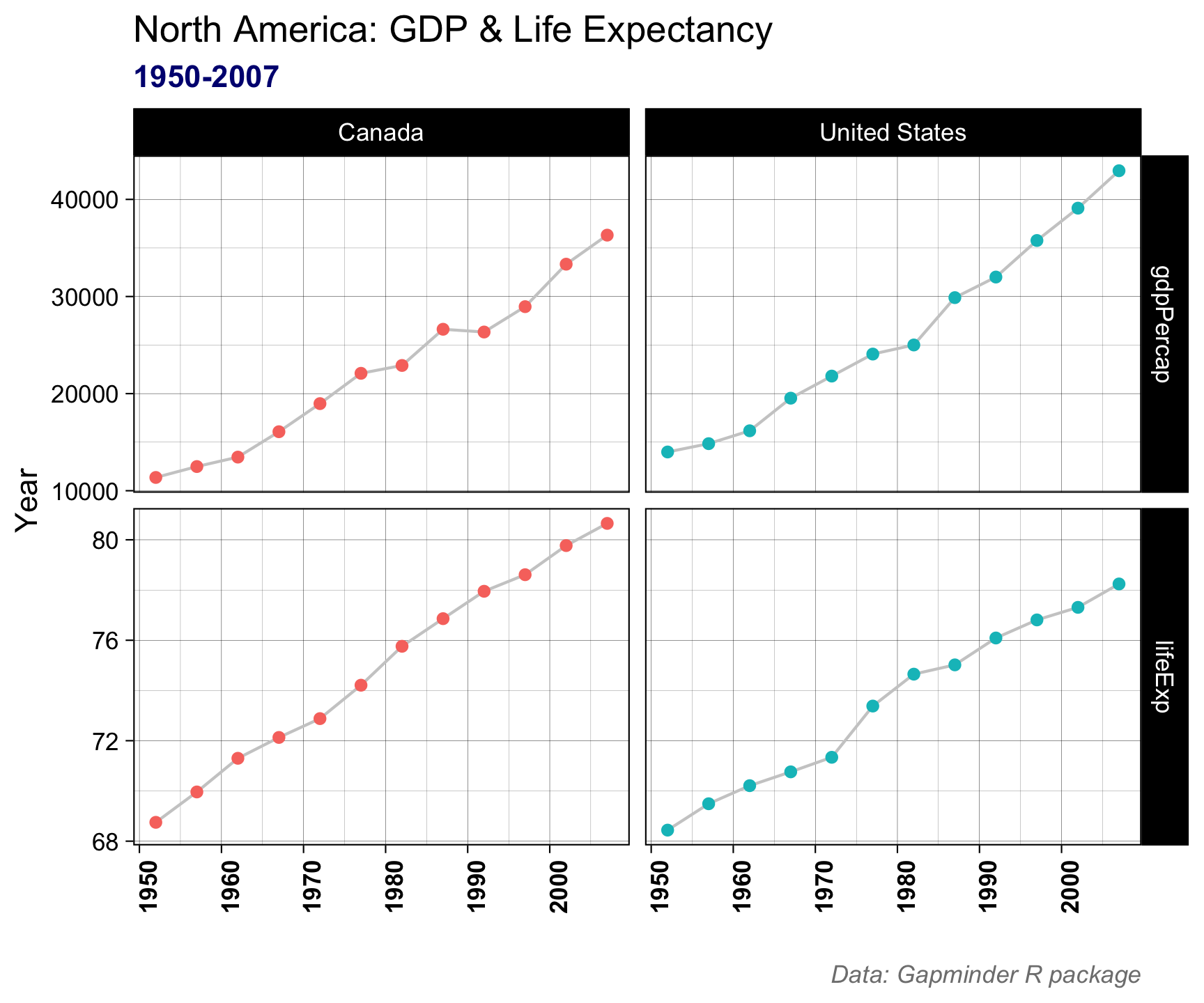
Summary
When to Use Long Data?
- Easier for aggregation, analysis, and plotting.
- Works well with ggplot2 and other tidyverse tools.
- Facilitates handling of missing values.
- Preferred for repeated measures or time series data.
When to Use Wide Data?
- Simpler for certain analyses (e.g., regression models).
- Easier to read when dealing with few variables.
- Can be easier for sharing datasets with non-technical audiences.
Assignment
In your ess-330-daily-exercises/R directory
- Create a new file called
day-08.R - Open that file.
- Add your name, date, and the purpose of the script as comments (preceded by #)
Assignment
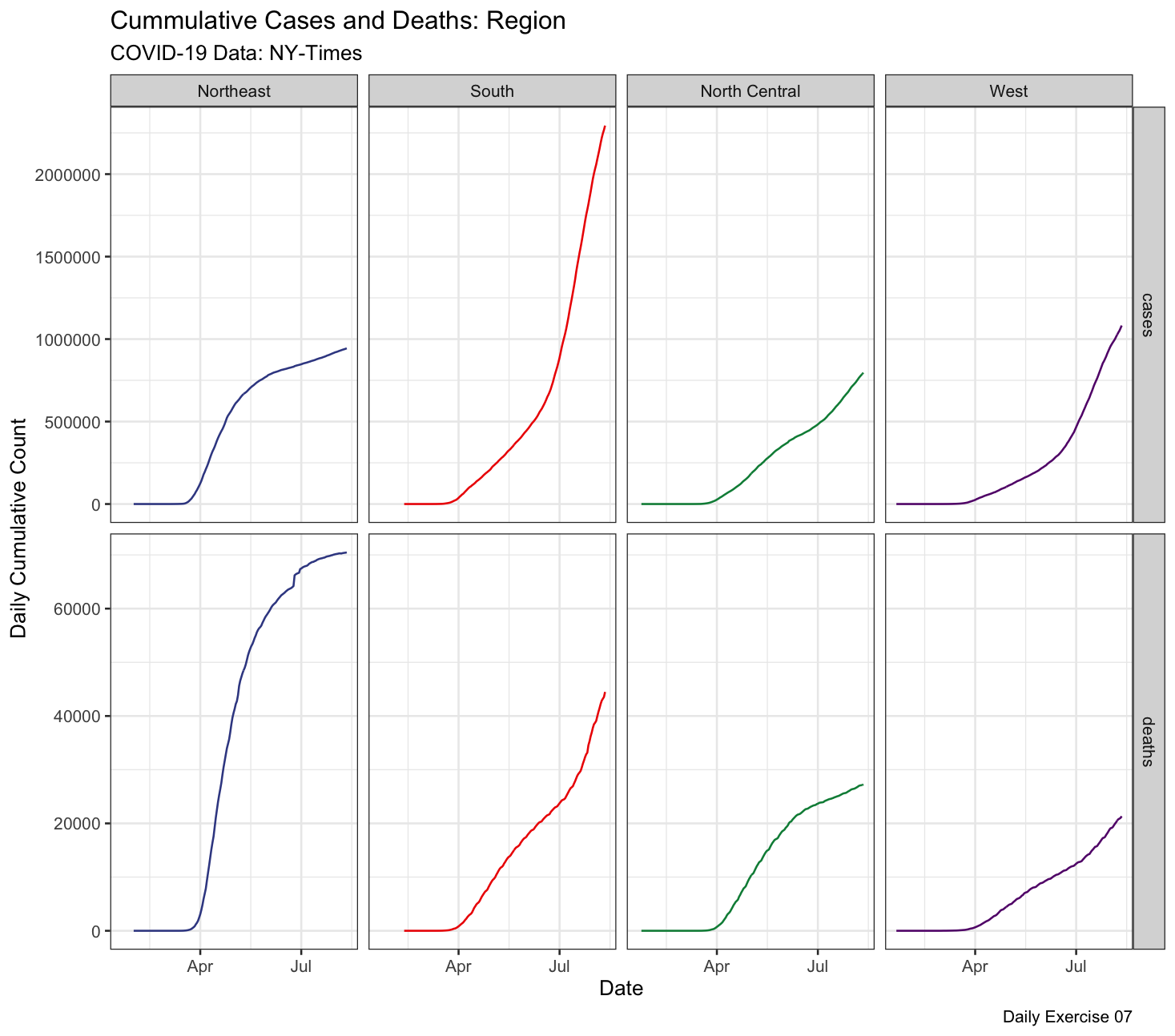
Question 1: Make a faceted plot of the cumulative cases & deaths by USA region. Your x axis should be the date and the y axis value/count. To do this you will need to join and pivot the COVID-19 data.
We can break this task into 7 steps:
- Read in the COVID-19 data
- Create a new data.frame using the available
state.abb,state.name,state.regionobjects in base R. Be intentional about creating a primary key to match to the COVID data!
- Join your new data.frame to the raw COVID data. Think about
right,inner,left, orfulljoin… - split-apply the joined data to determine the daily, cumulative, cases and deaths for each region
- Pivot your data from wide format to long
- Plot your data in a compelling way (setup, layers, labels, facets, themes)
- Save the image to your
imgdirectory with a good file name and extension!
Submission:
Push your work to Github
Turn in your Rscript, image, and repo URL to the Canvas dropbox