Lecture 02
The Digital Environment
2025-02-09
Slido!

Quantitative Reasoning

Working Definition:
Thinking about *data* from our world, to better understand and make choices about the past, present and future.Many people can work with data (e.g., data scientists), many people have domain knowledge (e.g., hydrologists), but few can do both in a careful way. That’s the goal of this class!
Almost all data is digital.
Working with computers is essential for quantitative analysis (precursor to reasoning).
Today is all about understanding how we interface with computers, their strengths, and limitations.
Your Environment
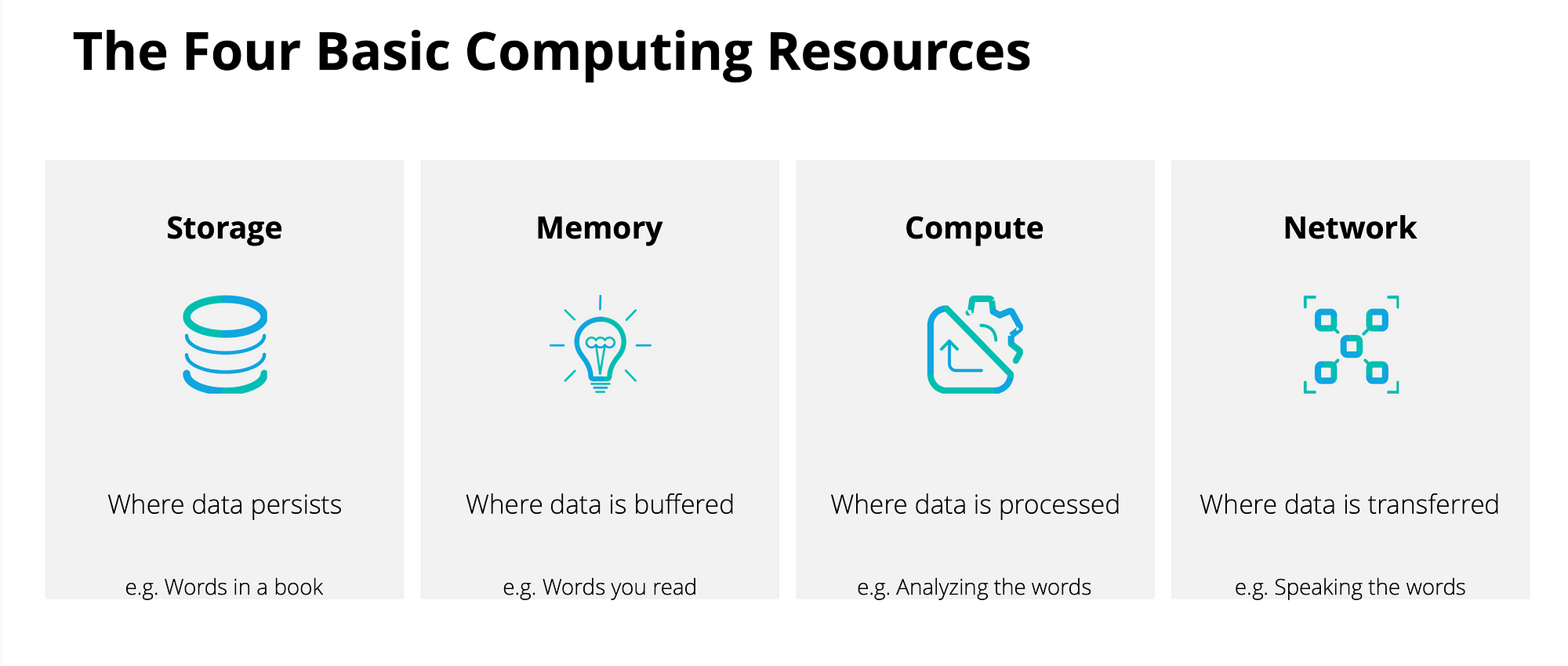
Computer: An electronic device for storing and processing data, typically in binary form, according to instruction. Computers store persistent data on disk, ephemeral data in memory, and executes processes with the CPU (Central Processing Unit).

File: A block of arbitrary information available to a computer program.
Compute
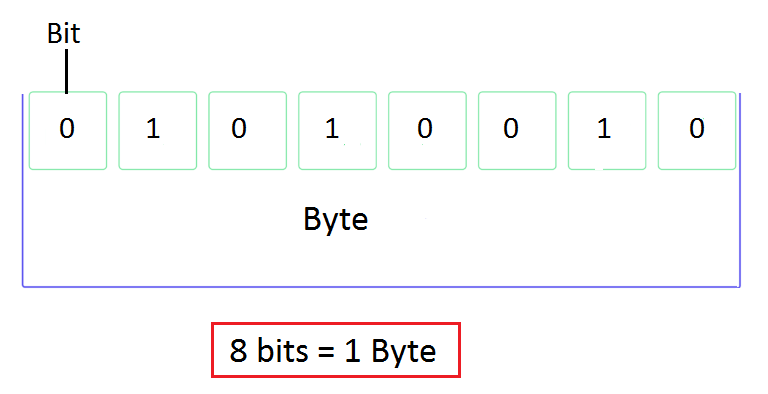
Bytes
Bytes vs Bits
A byte is a group of 8 binary digits (bits) and a unit of memory size.
The bit is a basic unit of information in computing and represents a logical state with two possible values (0 or 1).
Files
File Storage
- Files are stored as a collection of bytes on a hard drive
- Hard drives do not understand files - they just store bytes and directions to those bytes
- We need ways to retrieve (and write) bytes from (and to) the hard drive.
Interpreting Capacity

New computer capacity
Storage Pattern
File Storage
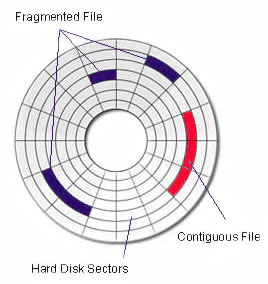
Defragmentation

What is a file?
Files save data on the hard drive as bytes in a meaningful way.
Files have three key properties:
- a name (machine and human interpret-able address)
- a path (a location in the file system)
- an extension (how to/what program reads the format)
Filesystem
Tip
For every file, there are paths and directories that lead to that specific file. These paths and directories are called File Systems.
Filesystem: describes the methods an operating system uses to organize files
If you are on a Windows device , and you want to find a file you just downloaded, you go to “This PC,” from where you click on “Documents” and there you find yet another folder called “Downloads” that has the downloaded files stored.
If you are on a Mac , to get the same result is click on “Downloads.” Whether you do it from the menu bar or anywhere else, it’s going to get you to exactly where you need to go.
Directories
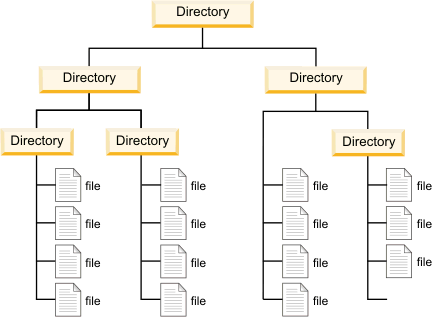
Directory: is a location for storing, organizing, and separating files and other directories on a computer. Think of folders!
Root directory: the “highest” or top-level directory in the hierarchy.
- The root directory contains all other folders/files in the drive or folder
- Sometimes referred to as the home directory
File Names
Tip
“There are only two hard things in Computer Science: cache invalidation and naming things.” ~Phil Karlton
- File names/paths are how we locate and identify information stored on a machine.
- Names are always up to us as users!
Three Principles for file names
machine readable
human readable
sortable
machine readable
avoid spaces, punctuation, accented characters, and mixed cases
regular expression friendly (e.g. use patterns!)
use ISO 8601 dates (YYYY-MM-DD)
be consistent with delimiters (easy to compute on)
Use “_” (underscores) to separate “metadata” you want latter
Use “-” (hyphens) to separate words for readability (like dates or names)
Doing so makes files …
easy to search for & narrow
#> [1] "1903-07-01_08033500_00060_tyler_tx.txt"
#> [2] "1923-10-01_08033000_00060_angelina_tx.txt"and metadata easy to recover (easy to compute on)
#> StartDate siteID parameterCode county state extension
#> 1 1903-07-01 08033500 00060 tyler tx txt
#> 2 1923-10-01 08033000 00060 angelina tx txt
#> 3 1923-10-01 08180500 00060 medina tx txt
#> 4 1923-12-01 08082500 00060 baylor tx txt
#> 5 1924-08-01 08062500 00060 ellis tx txthuman readable
- File names contain information about the content and purpose of the file
- easy to find the right file a year from now
- Here, we see the order that the files run (utilities, download, clean, analyze, figures), the project they belong to (src), are archived in the file names.
#> Order Project Purpose extension
#> 1 00 src utils R
#> 2 01 src data-download R
#> 3 02 src data-clean R
#> 4 03 src analysis R
#> 5 04 src figures RSort easily
put numeric values first (use leading 0 for 1-9)
use ISO 8601 dates (YYYY-MM-DD)
File Paths
File paths tell us the location of a file within the file system
Directories are stored as hierarchies, again with root (home) directory being the one holding everything on a system
The folder you are in, is called your working directory. (think
pwd)The folder above the working directory is the parent directory
All folders within the working directory are sub folders or child folder
Declaring File Paths
Files are located by their path. Think of this as the directions - from the root directory.
Directories are separated with backslashes (“\") on windows, and forward slashes (”/“) or MacOS and Linux machines.
Example:
/Users/mikejohnson/github/csu-ess-330/slides/images/03-bit-byte.png
- Root: /Users/mikejohnson
Absolute vs Realative Paths
There are two ways to specify a file path.
- An absolute path always begins in the root folder
- A relative path is relative to the current working directory
.. and . notation
The dot (.) and dot-dot (..) notation to help us write shorter paths
A single ‘.’ denotes “this directory”.
Two periods (“..”) means “the parent directory”

Enforcing Relative Paths
Tip
Working with absolute paths can be a pain compared to relative paths…
It is a good practice to keep all the files associated with a project — input data, R scripts, analytic results, figures - together.
This is such a common practice that RStudio has built-in support for this via projects.
A good project layout will ultimately make your life easier:
- It will help ensure the integrity of your data;
- It makes it simpler to share your code with someone else (a lab-mate, collaborator, or supervisor);
- It allows you to easily upload your code with your manuscript submission;
- It makes it easier to pick the project back up after a break.
File Extensions
All files store bits.
Extensions can be considered a type of metadata that provides information about the way data might be stored
There are 1000’s of different formats for data ranging from common to custom
Each format defines how the sequence of bits and bytes are laid out
Indicate the characteristics of the file, its intended use, and the default applications that can open/use the file.
If you double click a
.docxfile it opens in Word which interprets the meaning of the bytesIf you double click an
.Rfile it opens with RStudio, and R interprets the meaning of the bytes

Extension Interpretation
- Readers depended on anticipated structure
- The file is actually a PNG with the wrong file extension. “0x89 0x50” is how a PNG file starts.
- The data returned to R is a structured set of bits, interpreted according to the directions of the file and the interpreting language!
Bytes Being Interpreted
Bytes Being Interpreted
Bytes Being Interpreted
#> [1] "ESS 330"
#> [1] 45 53 53 20 33 33 30
#> [1] 01 00 01 00 00 00 01 00 01 01 00 00 01 00 01 00 01 01 00 00 01 00 01 00 00
#> [26] 00 00 00 00 01 00 00 01 01 00 00 01 01 00 00 01 01 00 00 01 01 00 00 00 00
#> [51] 00 00 01 01 00 00Bytes Being Interpreted
#> [1] "ESS 330"
#> [1] 45 53 53 20 33 33 30
#> [1] 01 00 01 00 00 00 01 00 01 01 00 00 01 00 01 00 01 01 00 00 01 00 01 00 00
#> [26] 00 00 00 00 01 00 00 01 01 00 00 01 01 00 00 01 01 00 00 01 01 00 00 00 00
#> [51] 00 00 01 01 00 00
#> [1] 56Bytes Being Interpreted
#> [1] "ESS 330"
#> [1] 45 53 53 20 33 33 30
#> [1] 01 00 01 00 00 00 01 00 01 01 00 00 01 00 01 00 01 01 00 00 01 00 01 00 00
#> [26] 00 00 00 00 01 00 00 01 01 00 00 01 01 00 00 01 01 00 00 01 01 00 00 00 00
#> [51] 00 00 01 01 00 00
#> [1] 56
#> [1] 7Bytes Being Interpreted
#> [1] "ESS 330"
#> [1] 45 53 53 20 33 33 30
#> [1] 01 00 01 00 00 00 01 00 01 01 00 00 01 00 01 00 01 01 00 00 01 00 01 00 00
#> [26] 00 00 00 00 01 00 00 01 01 00 00 01 01 00 00 01 01 00 00 01 01 00 00 00 00
#> [51] 00 00 01 01 00 00
#> [1] 56
#> [1] 7
#> [1] TRUEExample
Overview of File Types
- Text Files: Human-readable, easy to edit (e.g.,
.txt,.csv). - Binary Files: Optimized for performance, not human-readable (e.g.,
.nc,.tif). - Structured Files: Contain unified metadata and data (e.g.,
.json,.xlsx).
Practical Contexts for Files in Ecosystem Science
CSV: Used for tabular field survey data (e.g., species counts, water quality measurements).
GeoTIFF: Stores spatial data like satellite imagery or digital elevation models (DEMs).
NetCDF: Common for data with space and time data (multi-dimensional arrays) like climate models, hydrological simulations, and multidimensional data.
JSON/GeoJSON: Facilitates sharing geographic features in web applications or APIs.
GPKG (GeoPackage): An open, standards-based format for spatial data that supports vector and raster data.
SHP (Shapefile): Widely used for vector geographic data, though limited in attribute sizes and modern functionality.
Aside: URLs
Structure

- Protocol (Scheme): https, ftp, s3, …
- Subdomain
- Domain Name
- Top Level Domain (TLD)
- Path/File (w/ extension!)
- Parameters (APIs, databases, etc.)
Servers acts as a file system
https://mikejohnson51.github.io/
{Protocol (https)} / {subdomain} / {domain} / {top level domain}
https://mikejohnson51.github.io/csu-ess-330/
Protocol, subdomain, domain, TLD, path (repo)
https://mikejohnson51.github.io/csu-ess-330/schedule.html
Protocol, subdomain, domain, TLD, repo, file
https://mikejohnson51.github.io/csu-ess-330/slides/1-welcome.html#/title-slide
Protocol, subdomain, domain, TLD, repo, directory, file, component (html)
Cloud
s3://spatial-water-noaa/nwm/CONUS/ISLTYP.tif
{Protocol (s3)} / {bucket} / {directory} / {file}
Information
https://www.etix.com/ticket/p/53375671/steel-pulse-50th-anniversary-tour-fort-collins-washingtons
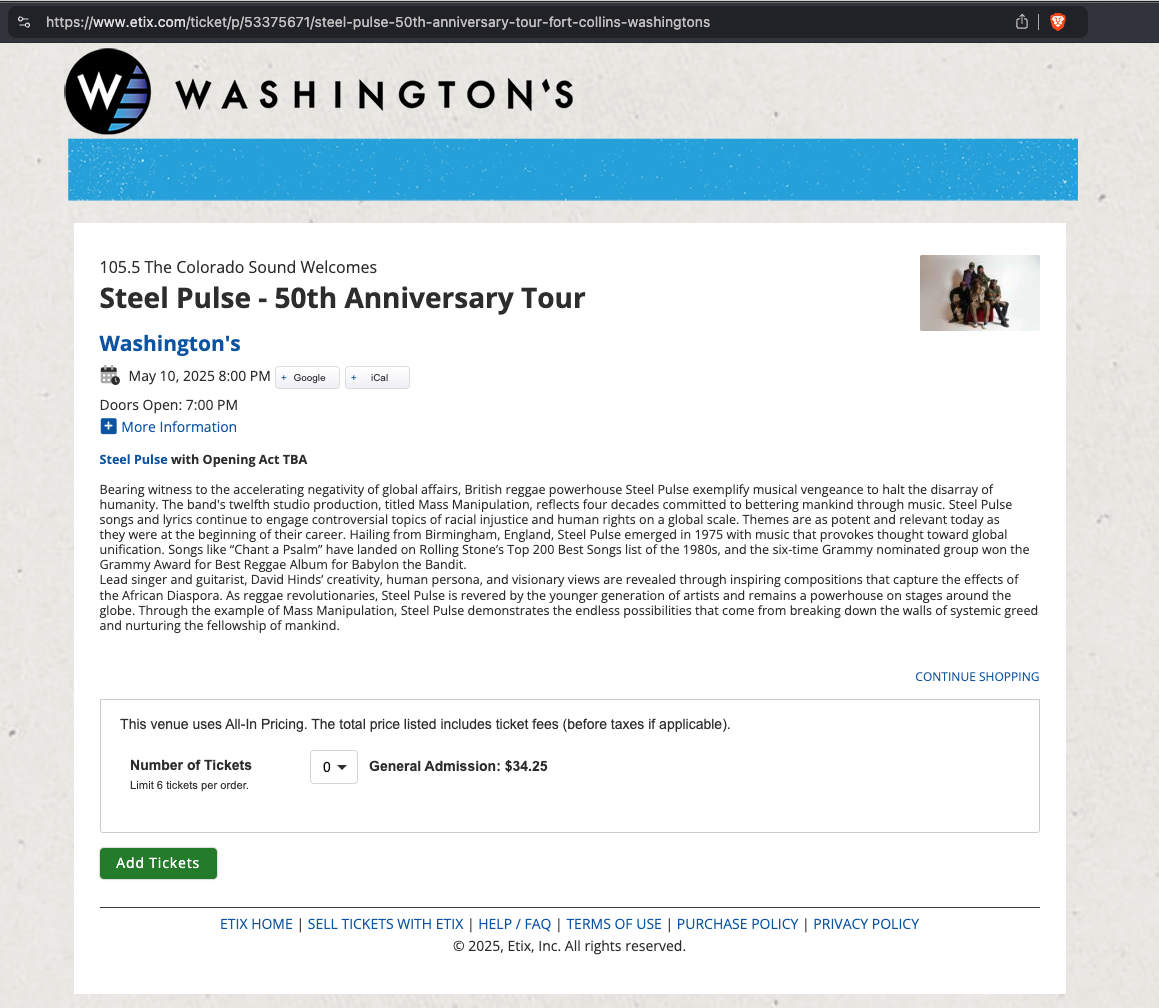
Important
index.html is everywhere online (lets take a look!)
Summary:

Next Time:
Daily Assignment: Set up Git and Github
Next Topic: Data Types

