Week 5
Machine Learning Part 1
Unit 3: Modeling (Machine Learning)

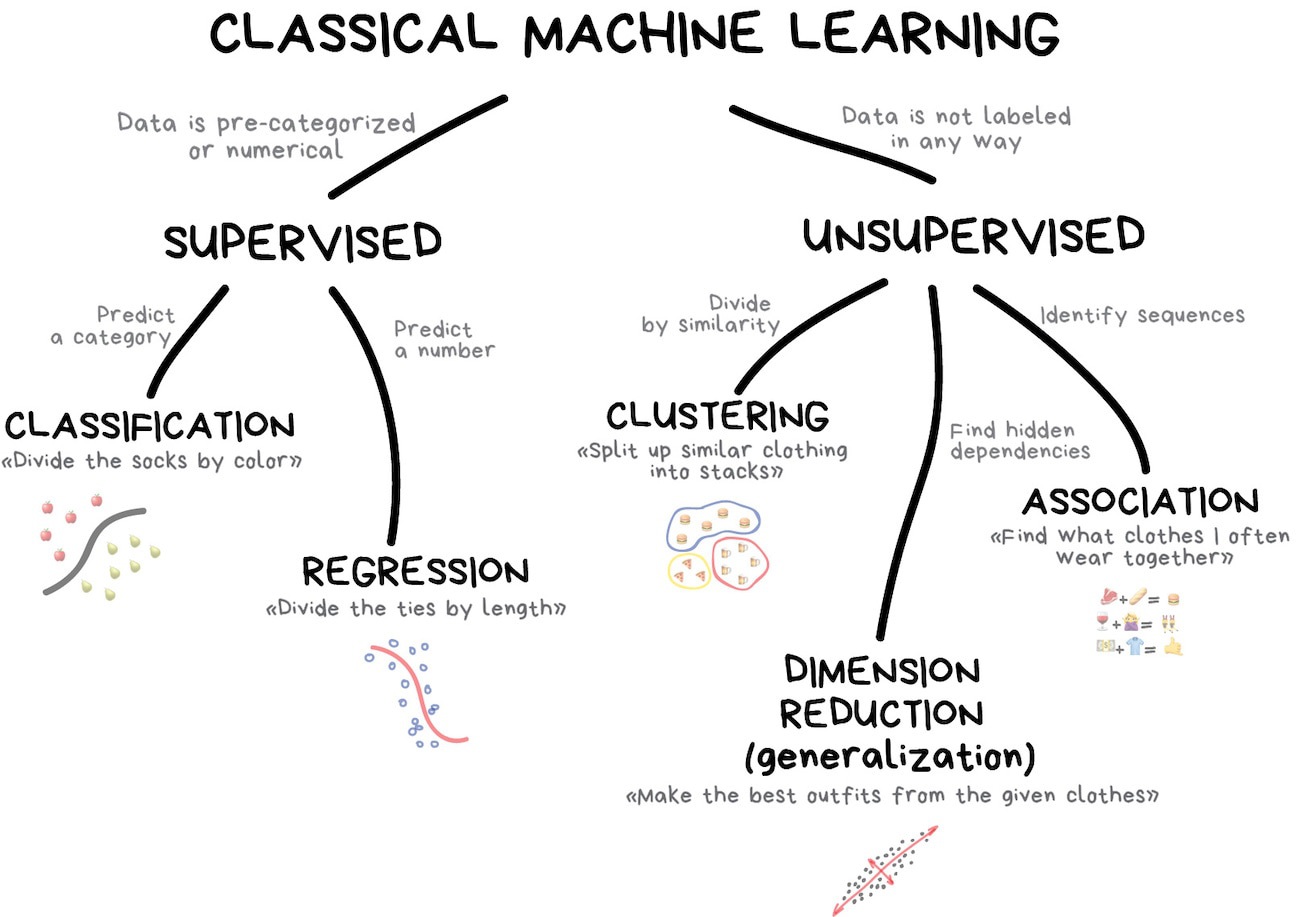
What is machine learning?
What is machine learning? (2025 edition)
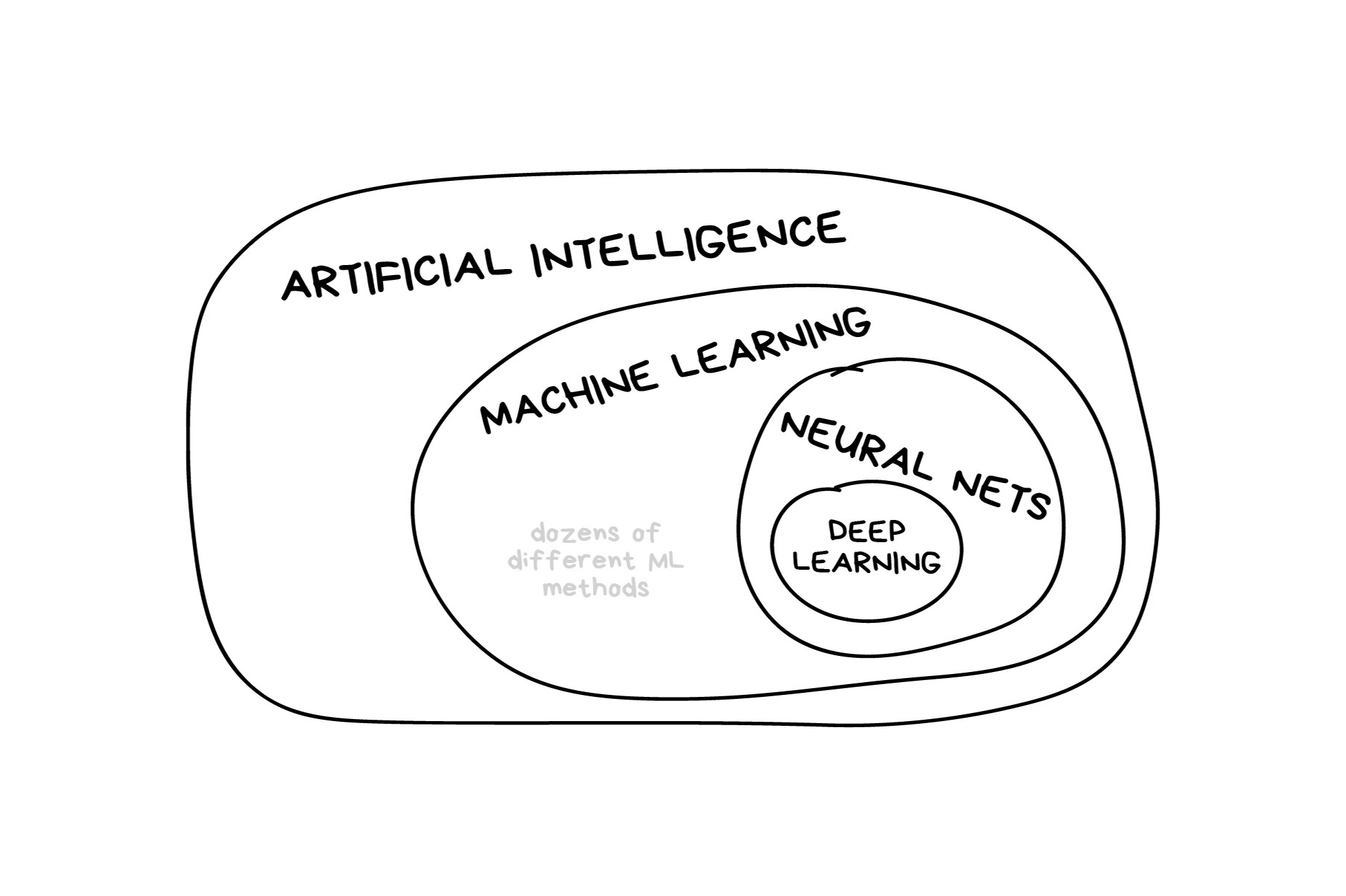
In the early 2010s, “Artificial intelligence” (AI) was largely synonymous with what we’ll refer to as “machine learning” in this workshop. In the late 2010s and early 2020s, AI usually referred to deep learning methods. Since the release of ChatGPT in late 2022, “AI” has come to also encompass large language models (LLMs) / generative models.
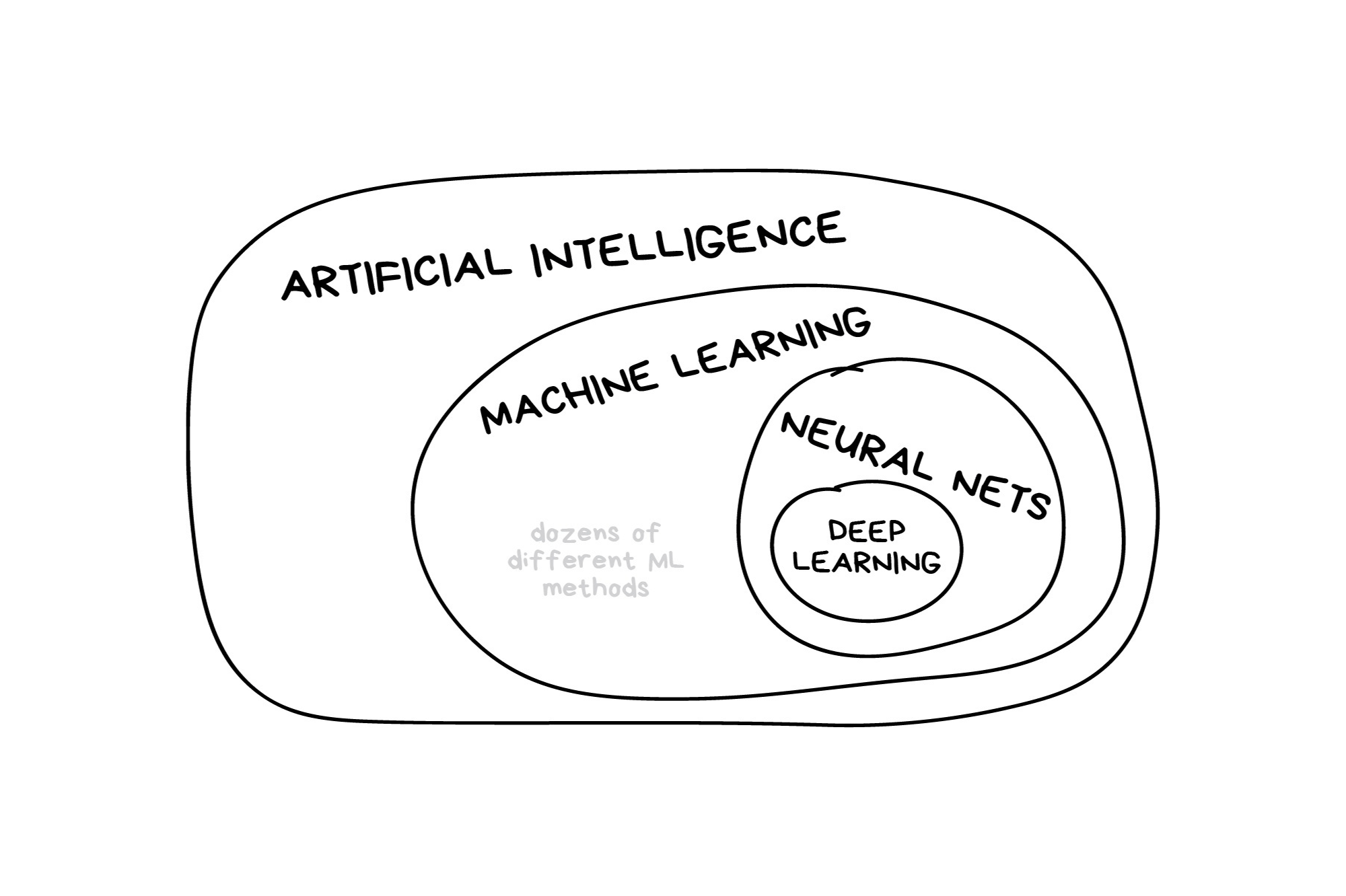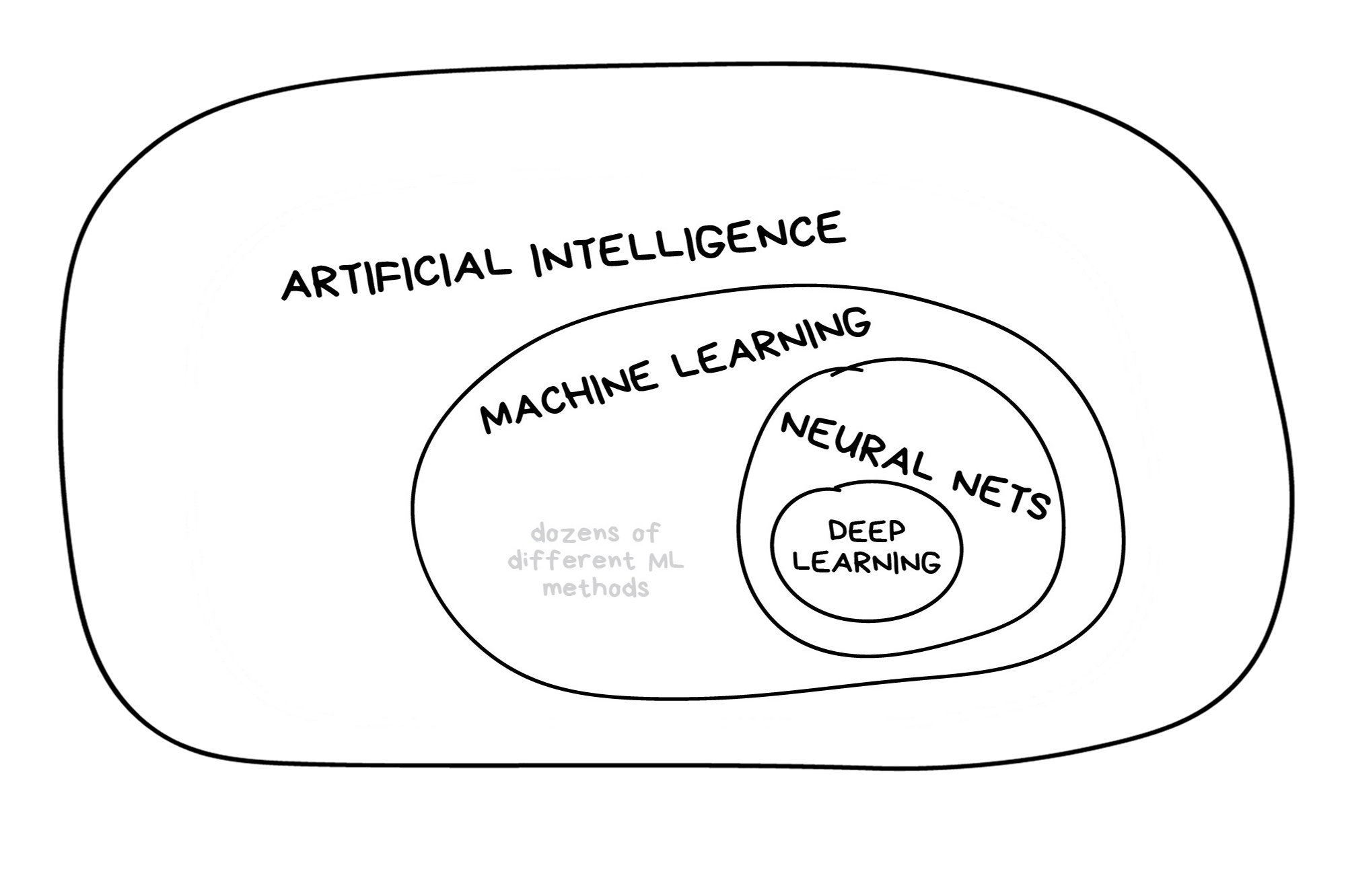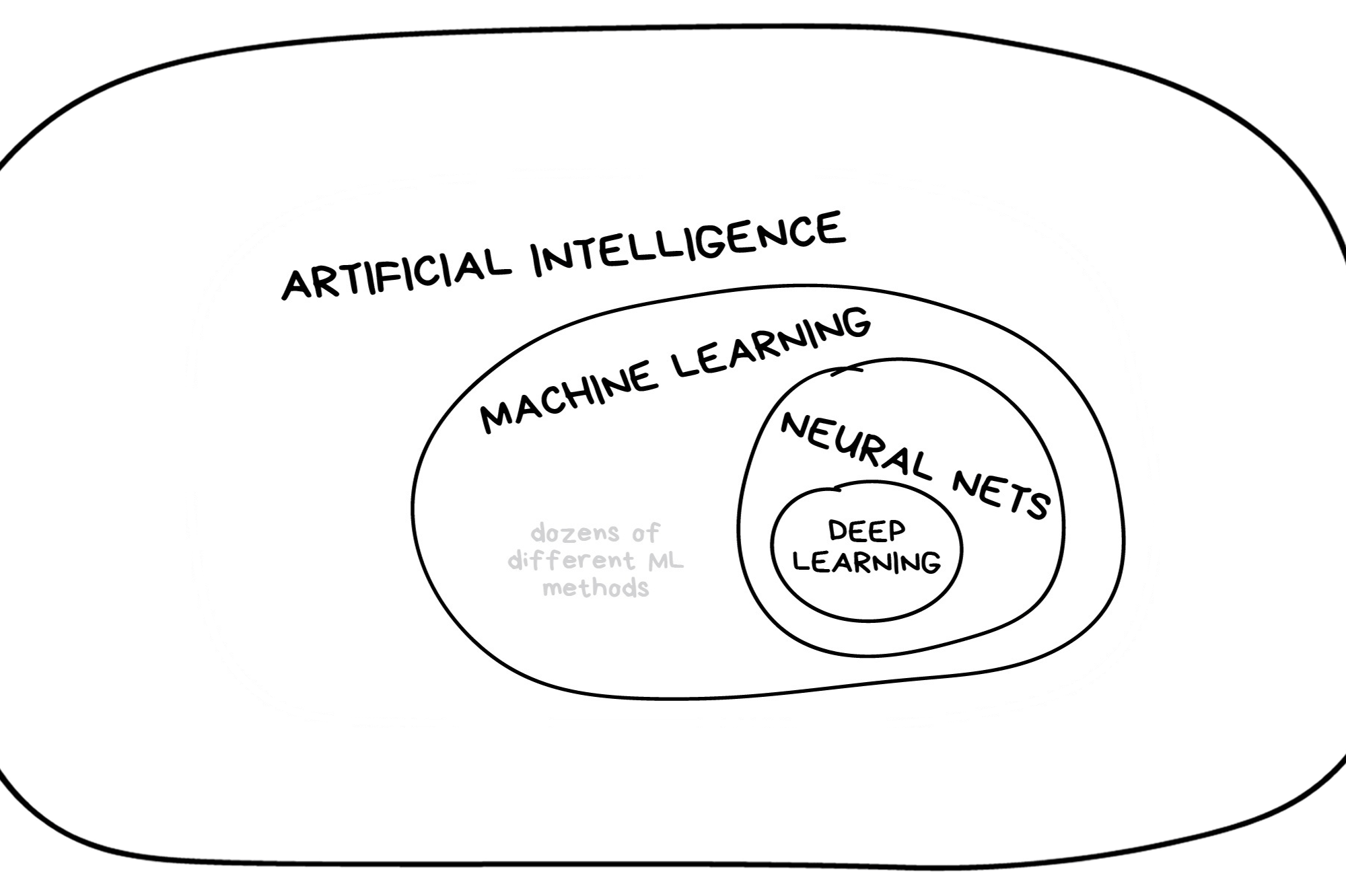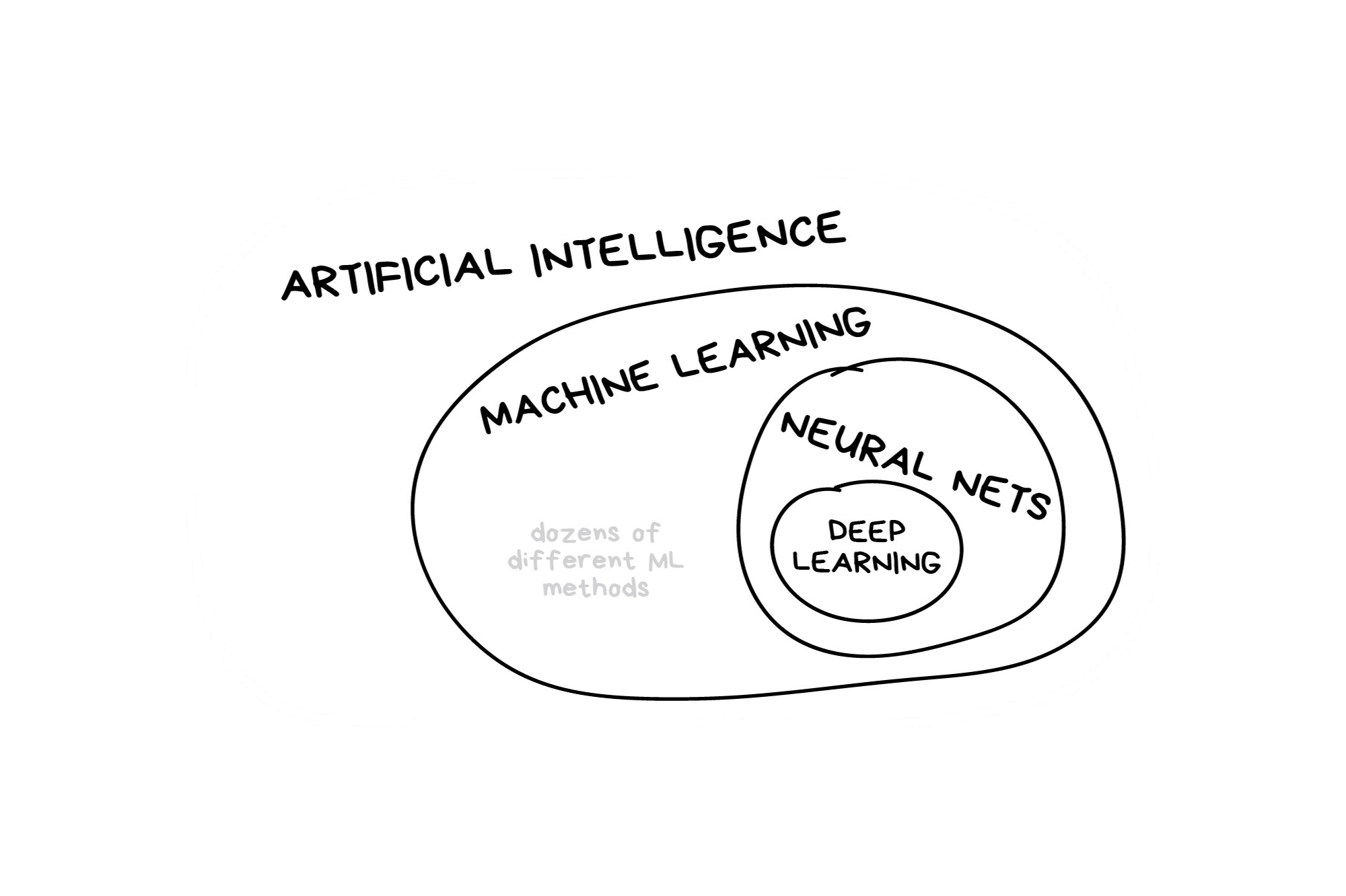
Classic Conceptual Model
The big picture: Road map
What is tidymodels?
- An R package ecosystem for modeling and machine learning
- Built on the
tidyverseprinciples (consistent syntax, modular design) - Provides a structured workflow for preprocessing, model fitting, and evaluation
- Includes packages for model specification, preprocessing, resampling, tuning, and evaluation
- Promotes best practices for reproducibility and efficiency
- Offers a unified interface for different models and tasks
Key tidymodels Packages
📦 recipes: Feature engineering and preprocessing
📦 parsnip: Unified interface for model specification
📦 workflows: Streamlined modeling pipelines
📦 tune: Hyperparameter tuning
📦 rsample: Resampling and validation
📦 yardstick: Model evaluation metrics
knitr::include_graphics('images/tidymodels-packages.png')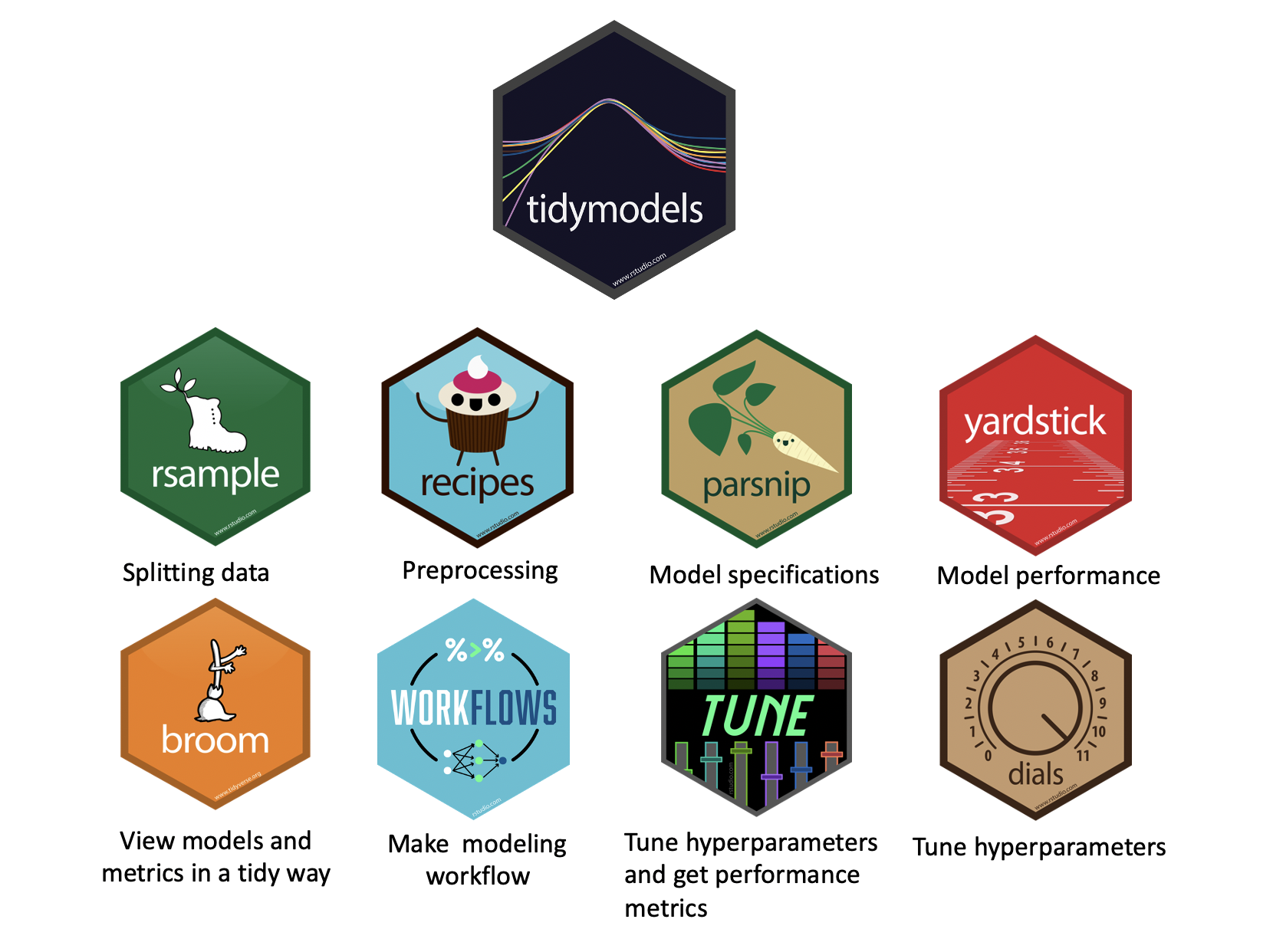
tidymodels::tidymodels_packages()
#> [1] "broom" "cli" "conflicted" "dials" "dplyr"
#> [6] "ggplot2" "hardhat" "infer" "modeldata" "parsnip"
#> [11] "purrr" "recipes" "rlang" "rsample" "rstudioapi"
#> [16] "tibble" "tidyr" "tune" "workflows" "workflowsets"
#> [21] "yardstick" "tidymodels"Typcical Workflow in tidymodels
1️⃣ Load Package & Data
2️⃣ Preprocess Data (recipes)
3️⃣ Define Model (parsnip)
4️⃣ Create a Workflow (workflows)
5️⃣ Train & Tune (tune)
6️⃣ Evaluate Performance (yardstick)
Data Normalization: tidymodels
Data normalization is a crucial step in data preprocessing for machine learning.
It ensures that features contribute equally to model performance by transforming them into a common scale.
This is particularly important for algorithms that rely on distance metrics (e.g., k-nearest neighbors, support vector machines) or gradient-based optimization (e.g., neural networks, logistic regression).
In the
tidymodelsframework, data normalization is handled within preprocessing workflows using therecipespackage, which provides a structured way to apply transformations consistently.
Why Normalize Data for ML?
✅ Improves Model Convergence: Many machine learning models rely on gradient descent, which can be inefficient if features are on different scales.
✅ Prevents Feature Domination: Features with large magnitudes can overshadow smaller ones, leading to biased models.
✅ Enhances Interpretability: Standardized data improves comparisons across variables and aids in better model understanding.
✅ Facilitates Distance-Based Methods: Algorithms like k-nearest neighbors and principal component analysis (PCA) perform best when data is normalized.
Common Normalization Techniques
1. Min-Max Scaling
Transforms data into a fixed range (typically [0,1]).
Preserves relationships but sensitive to outliers.
Formula: \[ x' = \frac{x - x_{min}}{x_{max} - x_{min}} \]
Implemented using
step_range()inrecipes.
2. Z-Score Standardization (Standard Scaling)
Centers data to have zero mean and unit variance.
More robust to outliers compared to min-max scaling.
Formula: \[ x' = \frac{x - \mu}{\sigma} \]
Implemented using
step_normalize()inrecipes.
Common Normalization Techniques
3. Robust Scaling
Uses median and interquartile range (IQR) for scaling.
Less sensitive to outliers.
Formula: \[ x' = \frac{x - median(x)}{IQR(x)} \]
Implemented using
step_YeoJohnson()orstep_BoxCox()inrecipes.
4. Log Transformation
Useful for skewed distributions to reduce the impact of extreme values.
Implemented using
step_log()inrecipes.
Common Normalization Techniques
5. Principal Component Analysis (PCA)
Projects data into a lower-dimensional space while maintaining variance.
Used in high-dimensional datasets.
Implemented using
step_pca()in recipes.
Other Feature Engineering Tasks
Handling Missing Data
step_impute_mean()step_impute_median()step_impute_knn()
can be used to fill missing values.
Encoding Categorical Variables
step_dummy()creates dummy variables for categorical features.step_other()groups infrequent categories into an “other” category.
Creating Interaction Terms
step_interact()generates interaction terms between predictors.
Feature Extraction
step_pca()orstep_ica()can be used to extract important components.
Text Feature Engineering
step_tokenize(), step_stopwords(), and step_tf() for text processing.
Binning Numerical Data
step_bin2factor() converts continuous variables into categorical bins.
Polynomial and Spline Features
step_poly() and step_bs() for generating polynomial and spline transformations.
Implementing Normalization in Tidymodels
What will we do?
We will create a recipe to normalize the numerical predictors in the
penguinsdataset using therecipespackage.We will then prepare (
prep) the recipe and apply it to the dataset to obtain normalized data.We will then
bakethe recipe to apply the transformations to the dataset.Once processed, we can implement a linear regression model using the normalized data.
. . .
Example Workflow
library(tidymodels)
(recipe_obj <- recipe(flipper_length_mm ~
bill_length_mm + body_mass_g +
sex + island + species,
data = penguins) |>
step_impute_mean(all_numeric_predictors()) |>
step_dummy(all_factor_predictors()) |>
step_normalize(all_numeric_predictors()))
# Prepare and apply transformations
prep_recipe <- prep(recipe_obj, training = penguins)
normalized_data <- bake(prep_recipe, new_data = NULL) |>
mutate(species = penguins$species) |>
drop_na()
head(normalized_data)
#> # A tibble: 6 × 9
#> bill_length_mm body_mass_g flipper_length_mm sex_male island_Dream
#> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 -0.886 -0.565 181 0.990 -0.750
#> 2 -0.812 -0.502 186 -1.01 -0.750
#> 3 -0.665 -1.19 195 -1.01 -0.750
#> 4 -1.33 -0.940 193 -1.01 -0.750
#> 5 -0.849 -0.690 190 0.990 -0.750
#> 6 -0.923 -0.721 181 -1.01 -0.750
#> # ℹ 4 more variables: island_Torgersen <dbl>, species_Chinstrap <dbl>,
#> # species_Gentoo <dbl>, species <fct>Explanation:
recipe()defines preprocessing steps for modeling.step_impute_mean(all_numeric_predictors())standardizes all numerical predictors.step_dummy(all_factor_predictors())creates dummy variables for categorical predictors.step_normalize(all_numeric_predictors()))standardizes all numerical predictors.prep()estimates parameters for transformations.bake()applies the transformations to the dataset.
Build a model with Normalized Data
(model = lm(flipper_length_mm ~ . , data = normalized_data) )#>
#> Call:
#> lm(formula = flipper_length_mm ~ ., data = normalized_data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm body_mass_g sex_male
#> 200.9649 2.2754 4.6414 0.7208
#> island_Dream island_Torgersen species_Chinstrap species_Gentoo
#> 0.6525 0.9800 0.5640 8.0796
#> speciesChinstrap speciesGentoo
#> NA NABuild a model with Normalized Data
(model = lm(flipper_length_mm ~ . , data = normalized_data) )
glance(model)#>
#> Call:
#> lm(formula = flipper_length_mm ~ ., data = normalized_data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm body_mass_g sex_male
#> 200.9649 2.2754 4.6414 0.7208
#> island_Dream island_Torgersen species_Chinstrap species_Gentoo
#> 0.6525 0.9800 0.5640 8.0796
#> speciesChinstrap speciesGentoo
#> NA NA
#> # A tibble: 1 × 12
#> r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0.863 0.861 5.23 294. 2.15e-136 7 -1020. 2057. 2092.
#> # ℹ 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>Build a model with Normalized Data
(model = lm(flipper_length_mm ~ . , data = normalized_data) )
glance(model)
(pred <- augment(model, normalized_data))#>
#> Call:
#> lm(formula = flipper_length_mm ~ ., data = normalized_data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm body_mass_g sex_male
#> 200.9649 2.2754 4.6414 0.7208
#> island_Dream island_Torgersen species_Chinstrap species_Gentoo
#> 0.6525 0.9800 0.5640 8.0796
#> speciesChinstrap speciesGentoo
#> NA NA
#> # A tibble: 1 × 12
#> r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0.863 0.861 5.23 294. 2.15e-136 7 -1020. 2057. 2092.
#> # ℹ 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>
#> # A tibble: 333 × 15
#> bill_length_mm body_mass_g flipper_length_mm sex_male island_Dream
#> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 -0.886 -0.565 181 0.990 -0.750
#> 2 -0.812 -0.502 186 -1.01 -0.750
#> 3 -0.665 -1.19 195 -1.01 -0.750
#> 4 -1.33 -0.940 193 -1.01 -0.750
#> 5 -0.849 -0.690 190 0.990 -0.750
#> 6 -0.923 -0.721 181 -1.01 -0.750
#> 7 -0.867 0.592 195 0.990 -0.750
#> 8 -0.518 -1.25 182 -1.01 -0.750
#> 9 -0.978 -0.502 191 0.990 -0.750
#> 10 -1.71 0.248 198 0.990 -0.750
#> # ℹ 323 more rows
#> # ℹ 10 more variables: island_Torgersen <dbl>, species_Chinstrap <dbl>,
#> # species_Gentoo <dbl>, species <fct>, .fitted <dbl>, .resid <dbl>,
#> # .hat <dbl>, .sigma <dbl>, .cooksd <dbl>, .std.resid <dbl>Build a model with Normalized Data
(model = lm(flipper_length_mm ~ . , data = normalized_data) )
glance(model)
(pred <- augment(model, normalized_data))
ggscatter(pred,
x = 'flipper_length_mm', y = '.fitted',
add = "reg.line", conf.int = TRUE,
cor.coef = TRUE, cor.method = "pearson",
color = "species", palette = "jco")#>
#> Call:
#> lm(formula = flipper_length_mm ~ ., data = normalized_data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm body_mass_g sex_male
#> 200.9649 2.2754 4.6414 0.7208
#> island_Dream island_Torgersen species_Chinstrap species_Gentoo
#> 0.6525 0.9800 0.5640 8.0796
#> speciesChinstrap speciesGentoo
#> NA NA
#> # A tibble: 1 × 12
#> r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0.863 0.861 5.23 294. 2.15e-136 7 -1020. 2057. 2092.
#> # ℹ 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>
#> # A tibble: 333 × 15
#> bill_length_mm body_mass_g flipper_length_mm sex_male island_Dream
#> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 -0.886 -0.565 181 0.990 -0.750
#> 2 -0.812 -0.502 186 -1.01 -0.750
#> 3 -0.665 -1.19 195 -1.01 -0.750
#> 4 -1.33 -0.940 193 -1.01 -0.750
#> 5 -0.849 -0.690 190 0.990 -0.750
#> 6 -0.923 -0.721 181 -1.01 -0.750
#> 7 -0.867 0.592 195 0.990 -0.750
#> 8 -0.518 -1.25 182 -1.01 -0.750
#> 9 -0.978 -0.502 191 0.990 -0.750
#> 10 -1.71 0.248 198 0.990 -0.750
#> # ℹ 323 more rows
#> # ℹ 10 more variables: island_Torgersen <dbl>, species_Chinstrap <dbl>,
#> # species_Gentoo <dbl>, species <fct>, .fitted <dbl>, .resid <dbl>,
#> # .hat <dbl>, .sigma <dbl>, .cooksd <dbl>, .std.resid <dbl>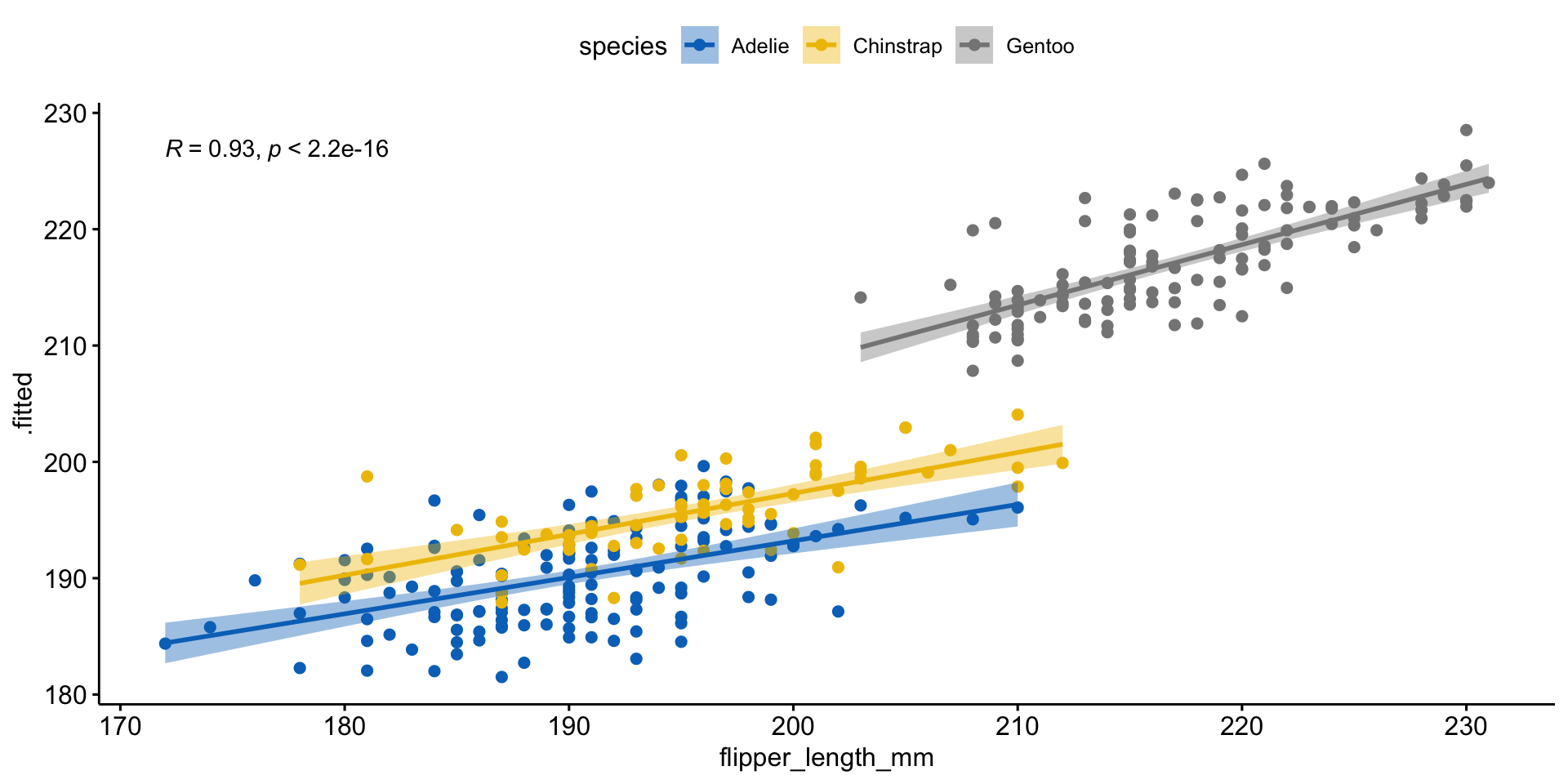
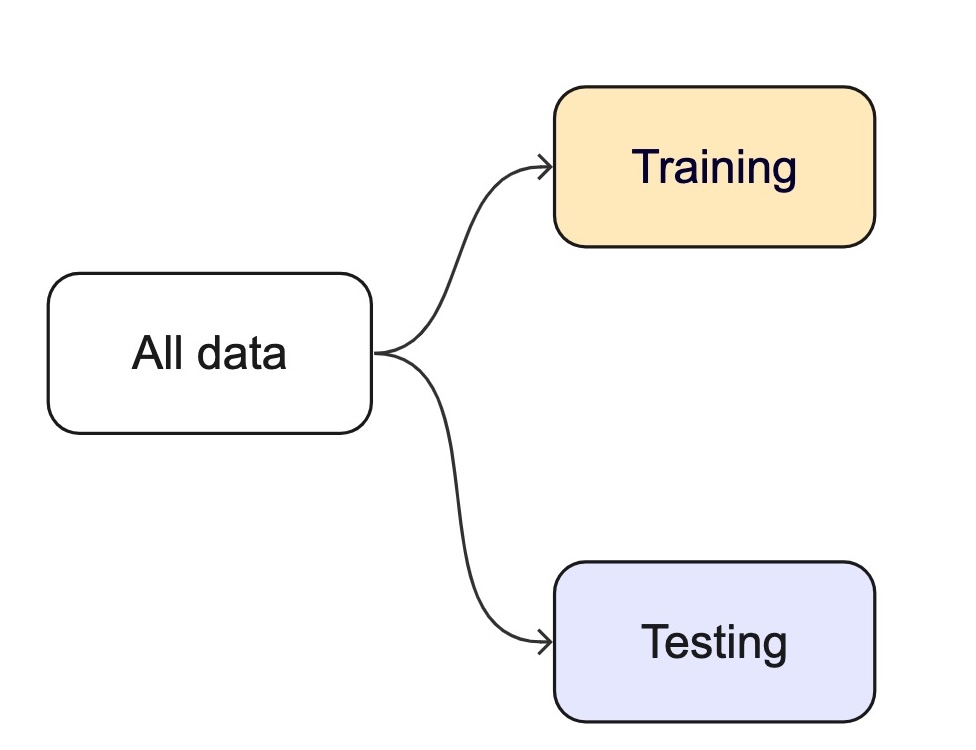
Data Budget
In any modeling effort, it’s crucial to evaluate the performance of a model using different validation techniques to ensure a model can generalize to unseen data.
But data is limited even in the age of “big data”.
Our typical process will like this:
- Read in raw data (
readr,dplyr::*_join) (single or multi table)
. . .
- Prepare the data (EDA/mutate/summarize/clean, etc)
- This is “Feature Engineering”
. . .
- Once we’ve established our features (rows), we decide how to “spend” them …
. . .
For machine learning, we typically split data into training and test sets:
. . .
The training set is used to estimate model parameters.
Spending too much data in training prevents us from computing a good assessment of model performance.
. . .
The test set is used as an independent assessment of performance.
Spending too much data in testing prevents us from computing good model parameter estimates.
How we split data

. . .
Splitting can be handled in many of ways. Typically, we base it off of a “hold out” percentage (e.g. 20%)
These hold out cases are extracted randomly from the data set. (remember seeds?)
. . .
- The training set is usually the majority of the data and provides a sandbox for testing different modeling appraoches.
. . .
The test set is held in reserve until one or two models are chosen.
The test set is then used as the final arbiter to determine the efficacy of the model.
Startification
In many cases, there is a structure to the data that would inhibit inpartial spliiting (e.g. a class, a region, a species, a sex, etc)
Imagine you have a jar of M&Ms in different colors — red, blue, and green. You want to take some candies out to taste, but you want to make sure you get a fair mix of each color, not just grabbing a bunch of red ones by accident.
Stratified resampling is like making sure that if your jar is 50% red, 30% blue, and 20% green, then the handful of candies you take keeps the same balance.
In data science, we do the same thing when we pick samples from a dataset: we make sure that different groups (like categories of people, animals, or weather types) are still fairly represented!
Initial splits 
- In
tidymodels, thersamplepackage provides functions for creating initial splits of data. - The
initial_split()function is used to create a single split of the data. - The
propargument defines the proportion of the data to be used for training. - The default is 0.75, which means 75% of the data will be used for training and 25% for testing.
set.seed(101991)
(resample_split <- initial_split(penguins, prop = 0.8))
#> <Training/Testing/Total>
#> <275/69/344>
#Sanity check
69/344
#> [1] 0.2005814Accessing the data: 
- Once the data is split, we can access the training and testing data using the
training()andtesting()functions to extract the partitioned data from the full set:
penguins_train <- training(resample_split)
glimpse(penguins_train)
#> Rows: 275
#> Columns: 7
#> $ species <fct> Gentoo, Adelie, Gentoo, Chinstrap, Gentoo, Chinstrap…
#> $ island <fct> Biscoe, Torgersen, Biscoe, Dream, Biscoe, Dream, Dre…
#> $ bill_length_mm <dbl> 46.2, 43.1, 46.2, 45.9, 46.5, 48.1, 45.2, 55.9, 38.1…
#> $ bill_depth_mm <dbl> 14.9, 19.2, 14.4, 17.1, 13.5, 16.4, 17.8, 17.0, 17.0…
#> $ flipper_length_mm <int> 221, 197, 214, 190, 210, 199, 198, 228, 181, 195, 20…
#> $ body_mass_g <int> 5300, 3500, 4650, 3575, 4550, 3325, 3950, 5600, 3175…
#> $ sex <fct> male, male, NA, female, female, female, female, male…. . .
penguins_test <- testing(resample_split)
glimpse(penguins_test)
#> Rows: 69
#> Columns: 7
#> $ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
#> $ island <fct> Torgersen, Torgersen, Torgersen, Biscoe, Biscoe, Bis…
#> $ bill_length_mm <dbl> NA, 39.3, 36.6, 35.9, 38.8, 37.9, 39.2, 39.6, 36.7, …
#> $ bill_depth_mm <dbl> NA, 20.6, 17.8, 19.2, 17.2, 18.6, 21.1, 17.2, 18.8, …
#> $ flipper_length_mm <int> NA, 190, 185, 189, 180, 172, 196, 196, 187, 205, 187…
#> $ body_mass_g <int> NA, 3650, 3700, 3800, 3800, 3150, 4150, 3550, 3800, …
#> $ sex <fct> NA, male, female, female, male, female, male, female…Proportional Gaps
# Dataset
(table(penguins$species) / nrow(penguins))
#>
#> Adelie Chinstrap Gentoo
#> 0.4418605 0.1976744 0.3604651
# Training Data
(table(penguins_train$species) / nrow(penguins_train))
#>
#> Adelie Chinstrap Gentoo
#> 0.4581818 0.1818182 0.3600000
# Testing Data
(table(penguins_test$species) / nrow(penguins_test))
#>
#> Adelie Chinstrap Gentoo
#> 0.3768116 0.2608696 0.3623188Stratification Example
A stratified random sample conducts a specified split within defined subsets of subsets, and then pools the results.
Only one column can be used to define a strata but grouping/mutate opperations can be used to create a new column that can be used as a strata (e.g. species/island)
In the case of the penguins data, we can stratify by species (think back to our nested/linear model appraoch)
This ensures that the training and testing sets have the same proportion of each species as the original data.
# Set the seed
set.seed(123)
# Drop missing values and split the data
penguins <- drop_na(penguins)
penguins_strata <- initial_split(penguins, strata = species, prop = .8)
# Extract the training and testing sets
train_strata <- training(penguins_strata)
test_strata <- testing(penguins_strata)Proportional Alignment
# Check the proportions
# Dataset
table(penguins$species) / nrow(penguins)
#>
#> Adelie Chinstrap Gentoo
#> 0.4384384 0.2042042 0.3573574
# Training Data
table(train_strata$species) / nrow(train_strata)
#>
#> Adelie Chinstrap Gentoo
#> 0.4377358 0.2037736 0.3584906
# Testing Data
table(test_strata$species) / nrow(test_strata)
#>
#> Adelie Chinstrap Gentoo
#> 0.4411765 0.2058824 0.3529412Modeling
Once the data is split, we can decide what type of model to invoke.
Often, users simply pick a well known model type or class for a type of problem (Classic Conceptual Model)
We will learn more about model types and uses next week!
For now, just know that the training data is used to fit a model.
If we are certain about the model, we can use the test data to evaluate the model.

Modeling Options
But, there are many types of models, with different assumptions, behaviors, and qualities, that make some more applicable to a given dataset!
Most models have some type of parmeterization, that can often be tuned.
Testing combinations of model and tuning parmaters also requires some combintation of
training/testingsplits.
Modeling Options
What if we want to compare more models?
. . .
And/or more model configurations?
. . .
And we want to understand if these are important differences?
. . .
How can we use the training data to compare and evaluate different models? 🤔
. . .
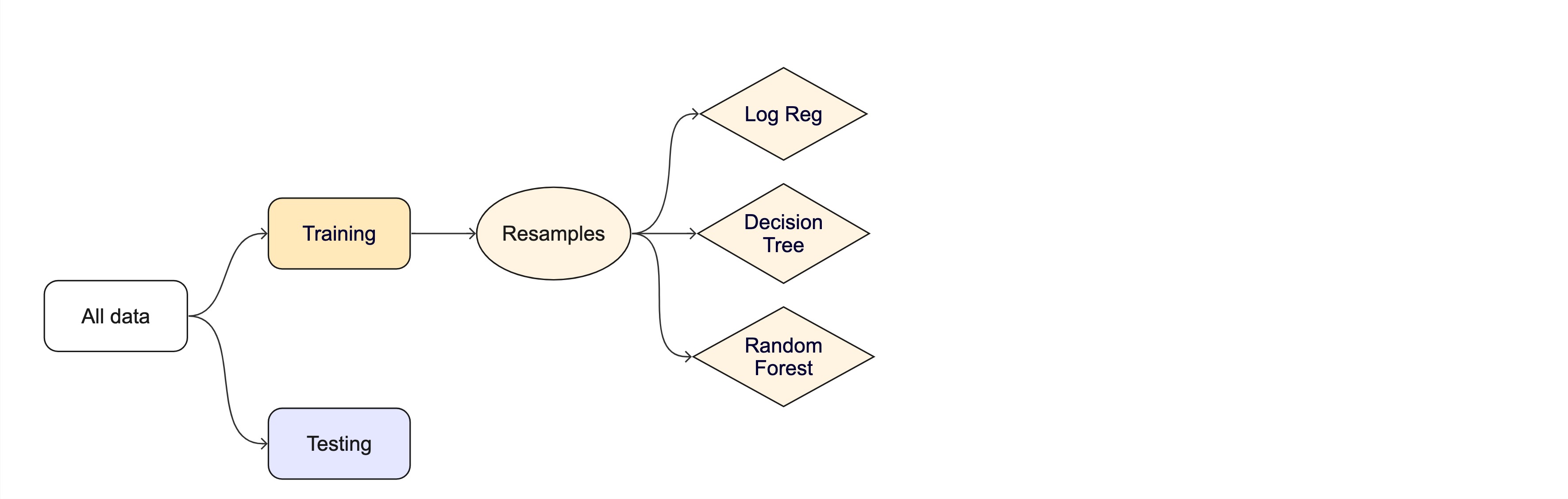
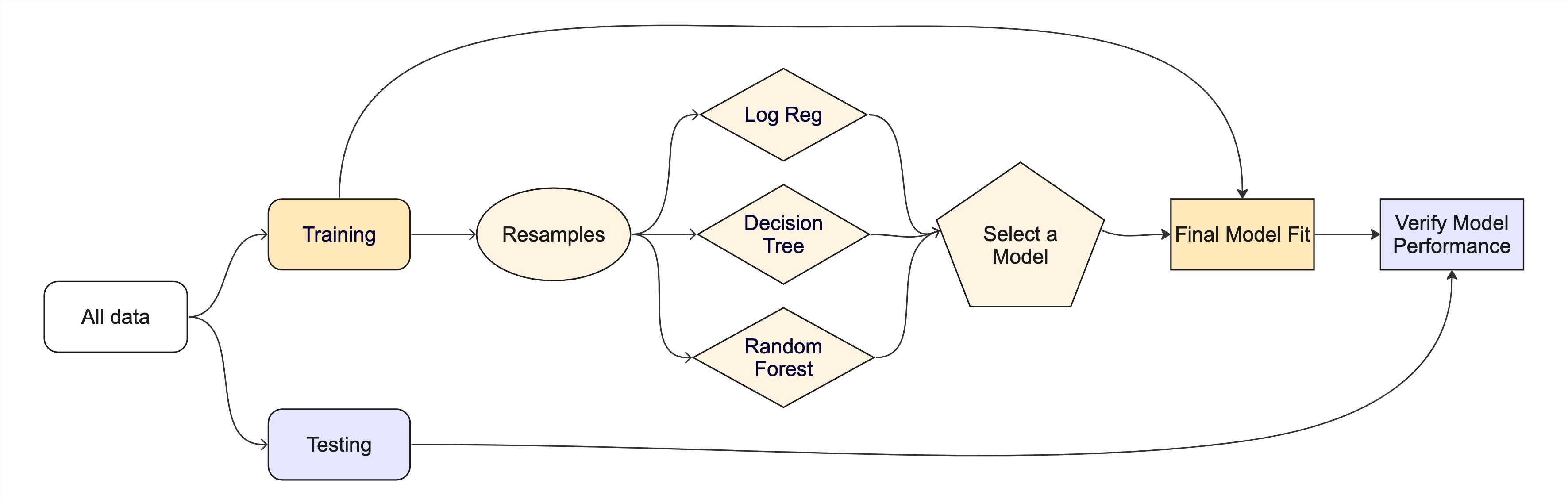
Resampling
Testing combinations of model and tuning parmaters also requires some combintation of
training/testingsplits.Resampling methods, such as cross-validation and bootstraping, are empirical simulation systems that help facilitate this.
They create a series of data sets similar to the initial
training/testingsplit.In the first level of the diagram to the right, we first split the original data into
training/testingsets. Then, the training set is chosen for resampling.
Key Resampling Methods
Resampling is always used with the training set.
Resampling is a key step in model validation and assessment. The rsample package provides tools to create resampling strategies such as cross-validation, bootstrapping, and validation splits.
. . .
- K-Fold Cross-Validation: The dataset is split into multiple “folds” (e.g., 5-fold or 10-fold), where each fold is used as a validation set while the remaining data is used for training.
. . .
- Bootstrap Resampling: Samples are drawn with replacement from the training dataset to generate multiple datasets, which used to train and test the model.
. . .
- Monte Carlo Resampling (Repeated Train-Test Splits): Randomly splits data multiple times.
. . .
- Validation Split: Creates a single training and testing partition.
Cross-validation
Cross-validation
Cross-validation 
penguins_train |> glimpse()
#> Rows: 275
#> Columns: 7
#> $ species <fct> Gentoo, Adelie, Gentoo, Chinstrap, Gentoo, Chinstrap…
#> $ island <fct> Biscoe, Torgersen, Biscoe, Dream, Biscoe, Dream, Dre…
#> $ bill_length_mm <dbl> 46.2, 43.1, 46.2, 45.9, 46.5, 48.1, 45.2, 55.9, 38.1…
#> $ bill_depth_mm <dbl> 14.9, 19.2, 14.4, 17.1, 13.5, 16.4, 17.8, 17.0, 17.0…
#> $ flipper_length_mm <int> 221, 197, 214, 190, 210, 199, 198, 228, 181, 195, 20…
#> $ body_mass_g <int> 5300, 3500, 4650, 3575, 4550, 3325, 3950, 5600, 3175…
#> $ sex <fct> male, male, NA, female, female, female, female, male…nrow(penguins_train) * 1/10
#> [1] 27.5
vfold_cv(penguins_train, v = 10) # v = 10 is default
#> # 10-fold cross-validation
#> # A tibble: 10 × 2
#> splits id
#> <list> <chr>
#> 1 <split [247/28]> Fold01
#> 2 <split [247/28]> Fold02
#> 3 <split [247/28]> Fold03
#> 4 <split [247/28]> Fold04
#> 5 <split [247/28]> Fold05
#> 6 <split [248/27]> Fold06
#> 7 <split [248/27]> Fold07
#> 8 <split [248/27]> Fold08
#> 9 <split [248/27]> Fold09
#> 10 <split [248/27]> Fold10Cross-validation 
What is in this?
penguins_folds <- vfold_cv(penguins_train)
penguins_folds$splits[1:3]
#> [[1]]
#> <Analysis/Assess/Total>
#> <247/28/275>
#>
#> [[2]]
#> <Analysis/Assess/Total>
#> <247/28/275>
#>
#> [[3]]
#> <Analysis/Assess/Total>
#> <247/28/275>Here is another example of a list column enabling the storage of non-atomic types in tibble
Set the seed when creating resamples
Alternate resampling schemes
Bootstrapping
Bootstrapping 
set.seed(3214)
bootstraps(penguins_train)
#> # Bootstrap sampling
#> # A tibble: 25 × 2
#> splits id
#> <list> <chr>
#> 1 <split [275/96]> Bootstrap01
#> 2 <split [275/105]> Bootstrap02
#> 3 <split [275/108]> Bootstrap03
#> 4 <split [275/111]> Bootstrap04
#> 5 <split [275/102]> Bootstrap05
#> 6 <split [275/98]> Bootstrap06
#> 7 <split [275/92]> Bootstrap07
#> 8 <split [275/97]> Bootstrap08
#> 9 <split [275/100]> Bootstrap09
#> 10 <split [275/99]> Bootstrap10
#> # ℹ 15 more rowsMonte Carlo Cross-Validation 
set.seed(322)
mc_cv(penguins_train, times = 10)
#> # Monte Carlo cross-validation (0.75/0.25) with 10 resamples
#> # A tibble: 10 × 2
#> splits id
#> <list> <chr>
#> 1 <split [206/69]> Resample01
#> 2 <split [206/69]> Resample02
#> 3 <split [206/69]> Resample03
#> 4 <split [206/69]> Resample04
#> 5 <split [206/69]> Resample05
#> 6 <split [206/69]> Resample06
#> 7 <split [206/69]> Resample07
#> 8 <split [206/69]> Resample08
#> 9 <split [206/69]> Resample09
#> 10 <split [206/69]> Resample10Validation set 
set.seed(853)
penguins_val_split <- initial_validation_split(penguins)
penguins_val_split
#> <Training/Validation/Testing/Total>
#> <199/67/67/333>
validation_set(penguins_val_split)
#> # A tibble: 1 × 2
#> splits id
#> <list> <chr>
#> 1 <split [199/67]> validation. . .
A validation set is just another type of resample
The whole game - status update
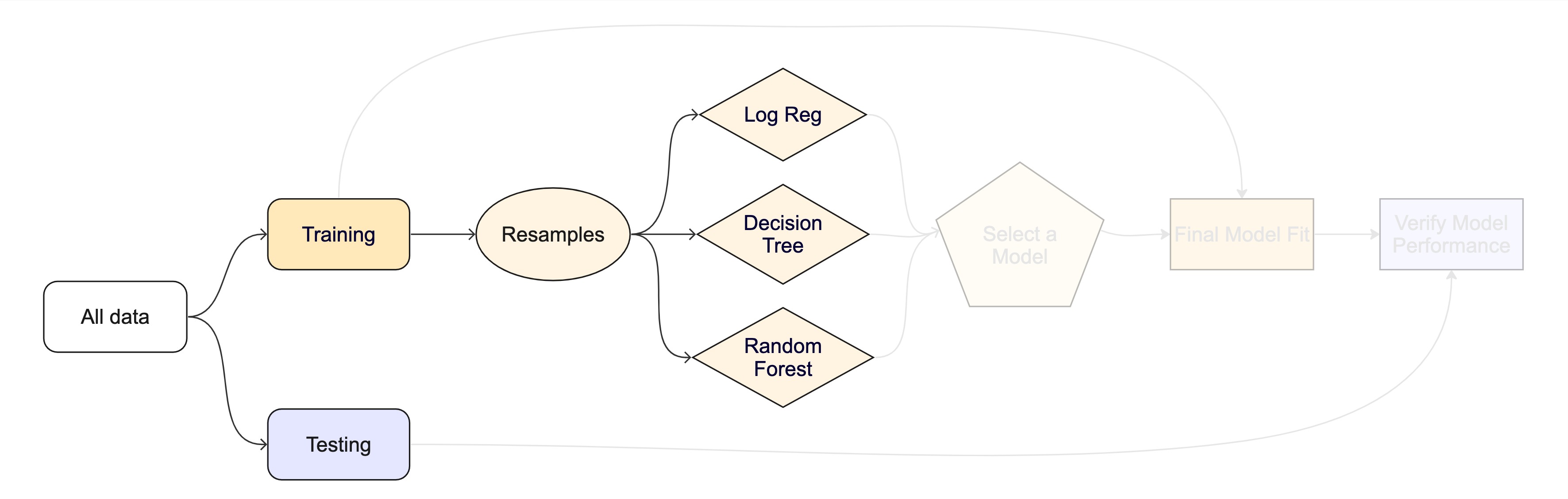
Unit Example
Can we predict if a plot of land is forested? - Interested in classification models - We have a dataset of 7,107 6000-acre hexagons in Washington state. - Each hexagon has a nominal outcome, forested, with levels "Yes" and "No".
Data on forests in Washington
- The U.S. Forest Service maintains ML models to predict whether a plot of land is “forested.”
- This classification is important for all sorts of research, legislation, and land management purposes.
- Plots are typically remeasured every 10 years and this dataset contains the most recent measurement per plot.
- Type
?forestedto learn more about this dataset, including references.
Data on forests in Washington
library(tidymodels)
library(forested)
dim(forested)
#> [1] 7107 19One observation from each of 7,107 6000-acre hexagons in Washington state.
A nominal outcome,
forested, with levels"Yes"and"No", measured “on-the-ground.”
table(forested$forested)
#>
#> Yes No
#> 3894 3213- 18 remotely-sensed and easily-accessible predictors:
names(forested)
#> [1] "forested" "year" "elevation" "eastness"
#> [5] "northness" "roughness" "tree_no_tree" "dew_temp"
#> [9] "precip_annual" "temp_annual_mean" "temp_annual_min" "temp_annual_max"
#> [13] "temp_january_min" "vapor_min" "vapor_max" "canopy_cover"
#> [17] "lon" "lat" "land_type"Data splitting and spending
The initial split 
set.seed(123)
forested_split <- initial_split(forested, prop = .8, strata = tree_no_tree)
forested_split
#> <Training/Testing/Total>
#> <5685/1422/7107>Accessing the data 
forested_train <- training(forested_split)
forested_test <- testing(forested_split)
nrow(forested_train)
#> [1] 5685
nrow(forested_test)
#> [1] 1422K-Fold Cross-Validation 
# Load the dataset
set.seed(123)
# Create a 10-fold cross-validation object
(forested_folds <- vfold_cv(forested_train, v = 10))
#> # 10-fold cross-validation
#> # A tibble: 10 × 2
#> splits id
#> <list> <chr>
#> 1 <split [5116/569]> Fold01
#> 2 <split [5116/569]> Fold02
#> 3 <split [5116/569]> Fold03
#> 4 <split [5116/569]> Fold04
#> 5 <split [5116/569]> Fold05
#> 6 <split [5117/568]> Fold06
#> 7 <split [5117/568]> Fold07
#> 8 <split [5117/568]> Fold08
#> 9 <split [5117/568]> Fold09
#> 10 <split [5117/568]> Fold10How do you fit a linear model in R?
lmfor linear model <– The one we have looked at!glmnetfor regularized regressionkerasfor regression using TensorFlowstanfor Bayesian regression using Stansparkfor large data sets using sparkbruleefor regression using torch (PyTourch)
Challenge
- All of these models have different syntax and functions
- How do you keep track of all of them?
- How do you know which one to use?
- How would you compare them?
Comparing 3-5 of these models is a lot of work using functions for diverse packages.
The tidymodels advantage
- In the
tidymodelsframework, all models are created using the same syntax:- This makes it easy to compare models
- This makes it easy to switch between models
- This makes it easy to use the same model with different engines (packages)
- The
parsnippackage provides a consistent interface to many models - For example, to fit a linear model you would be able to access the
linear_reg() function
?linear_reg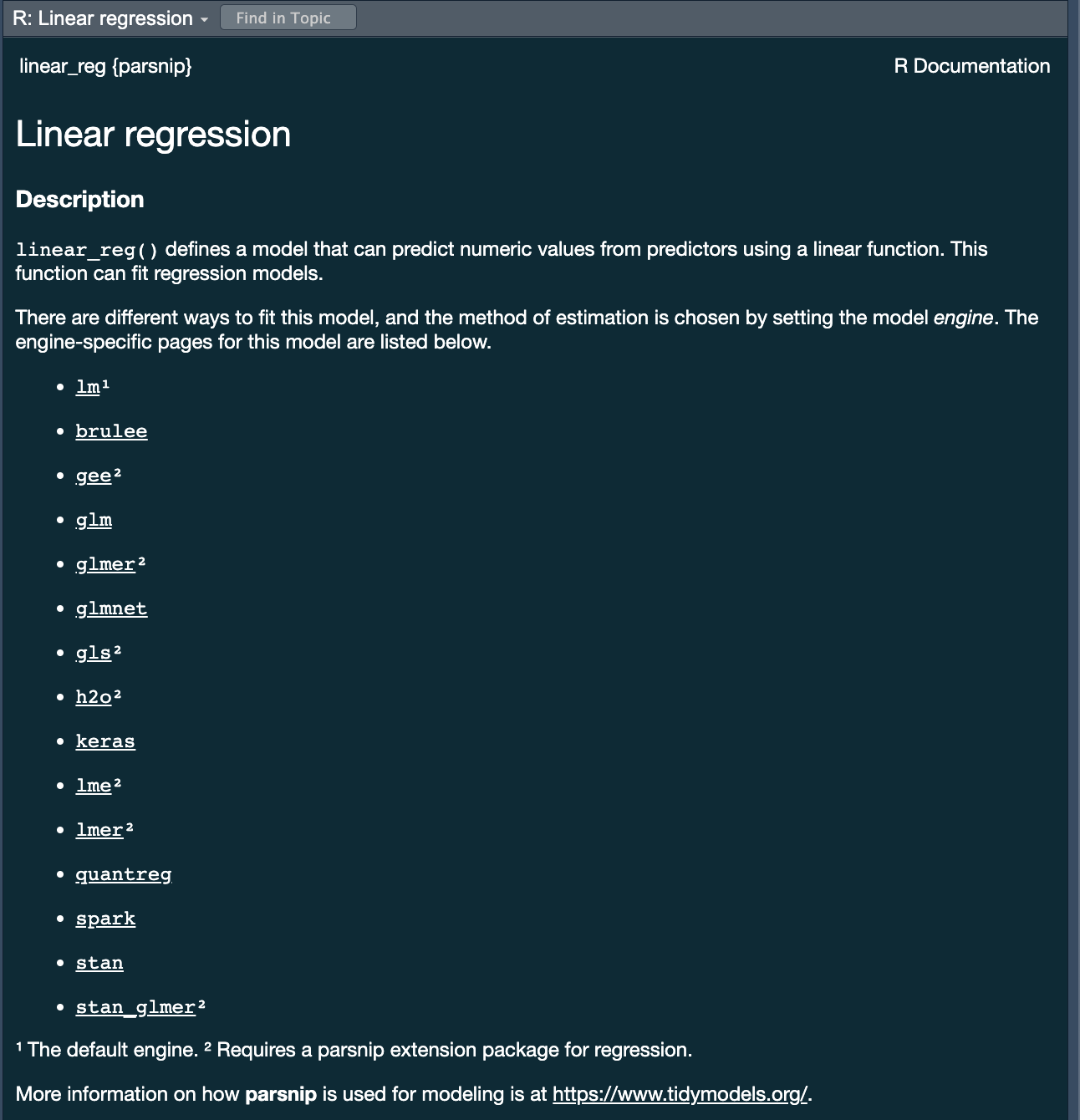
A tidymodels prediction will … 
- always be inside a tibble
- have column names and types are unsurprising and predictable
- ensure the number of rows in
new_dataand the output are the same
To specify a model 
. . .
- Choose a model
- Specify an engine
- Set the mode
Specify a model: Type 
Next week we will discuss model types more thoroughly. For now, we will focus on two types:
- Logistic Regression
logistic_reg()
#> Logistic Regression Model Specification (classification)
#>
#> Computational engine: glmdecision_tree()
#> Decision Tree Model Specification (unknown mode)
#>
#> Computational engine: rpartWe chose these two because they are robust, simple model that fit our goal of predicting a binary condition/class (forested/not forested)
Specify a model: engine 
- Choose a model
- Specify an engine
- Set the mode
?logistic_reg()
Specify a model: engine 
logistic_reg() %>%
set_engine("glmnet")
#> Logistic Regression Model Specification (classification)
#>
#> Computational engine: glmnet. . .
logistic_reg() %>%
set_engine("stan")
#> Logistic Regression Model Specification (classification)
#>
#> Computational engine: stanSpecify a model: mode 
- Choose a model
- Specify an engine
- Set the mode
?logistic_reg()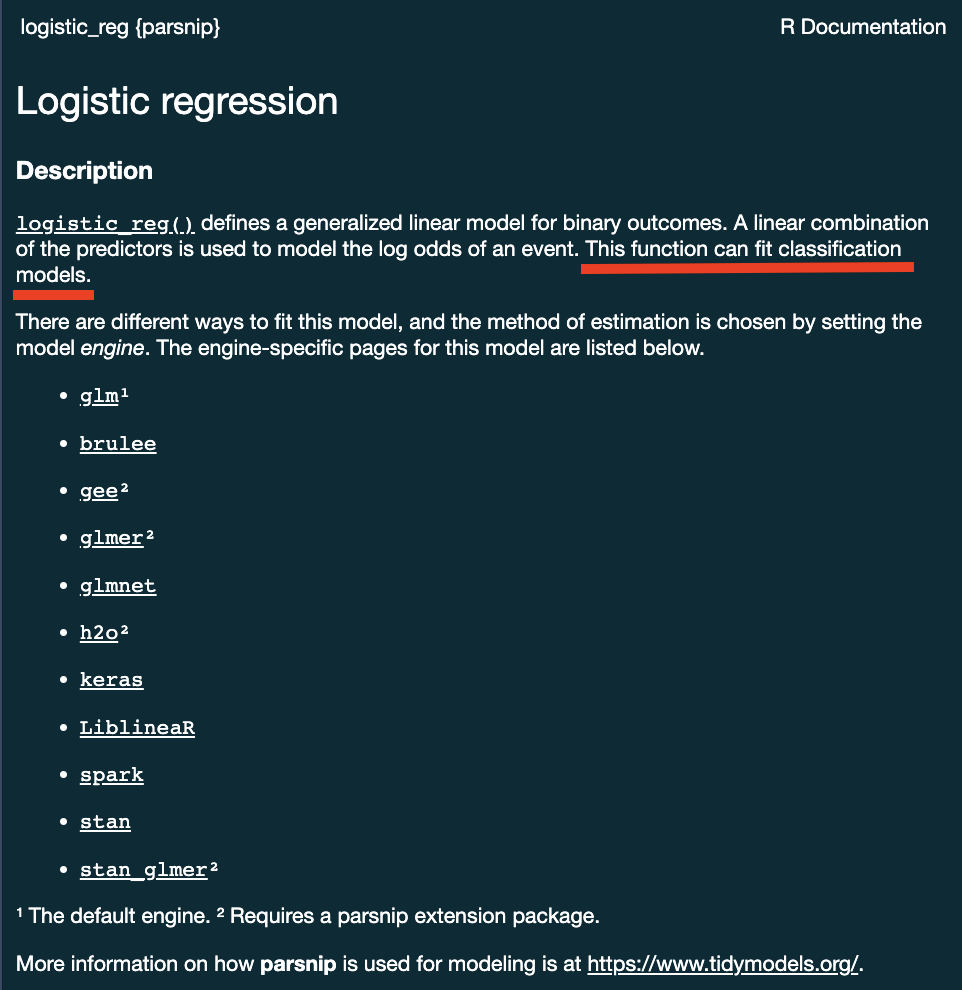
Specify a model: mode 
Some models have limit classes …
logistic_reg()
#> Logistic Regression Model Specification (classification)
#>
#> Computational engine: glm. . .
Others requires specification …
decision_tree()
#> Decision Tree Model Specification (unknown mode)
#>
#> Computational engine: rpart
decision_tree() %>%
set_mode("classification")
#> Decision Tree Model Specification (classification)
#>
#> Computational engine: rpartAll available models are listed at https://www.tidymodels.org/find/parsnip/
To specify a model 
- Choose a model
- Specify an engine
- Set the mode
Logistic regression
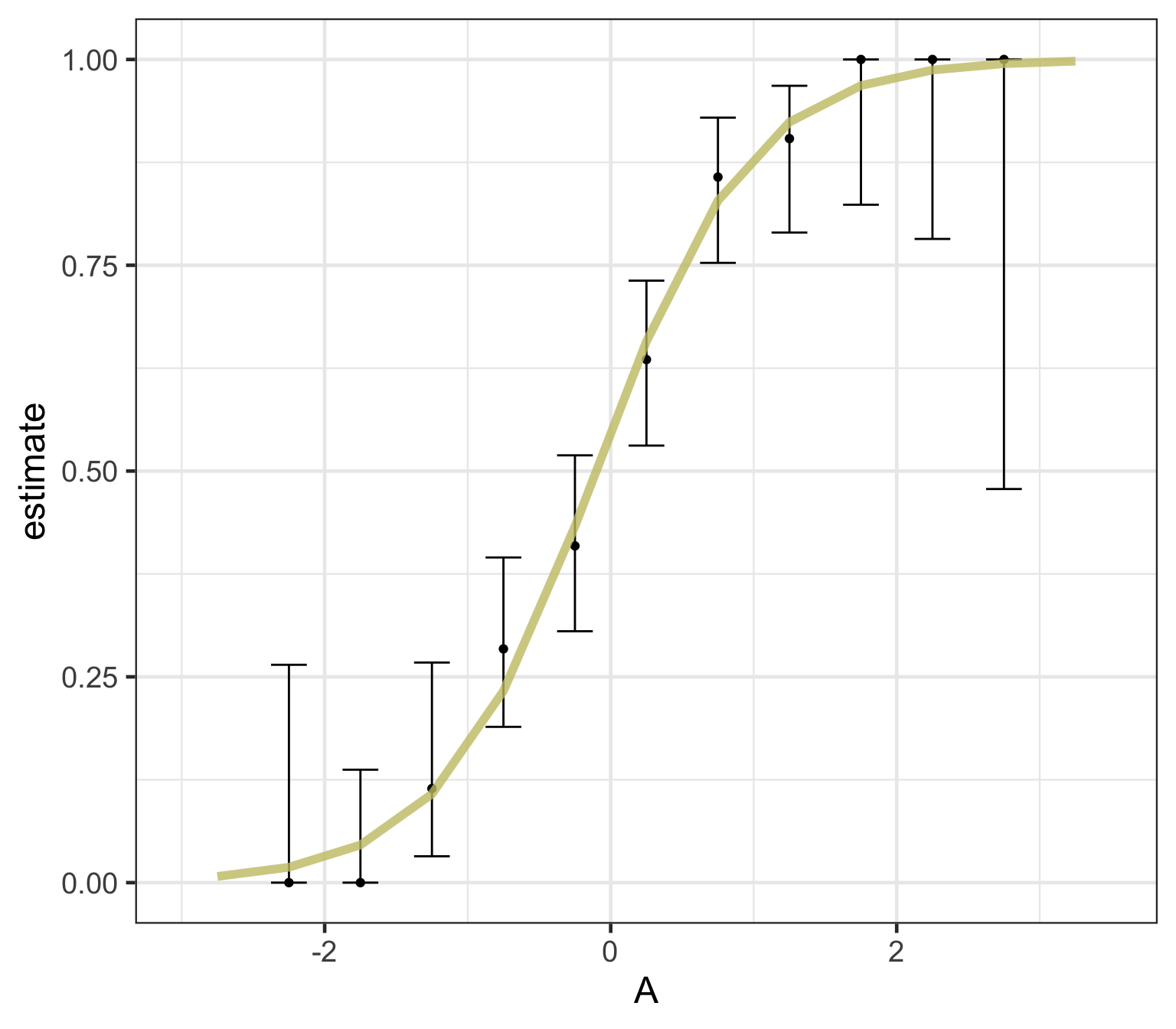
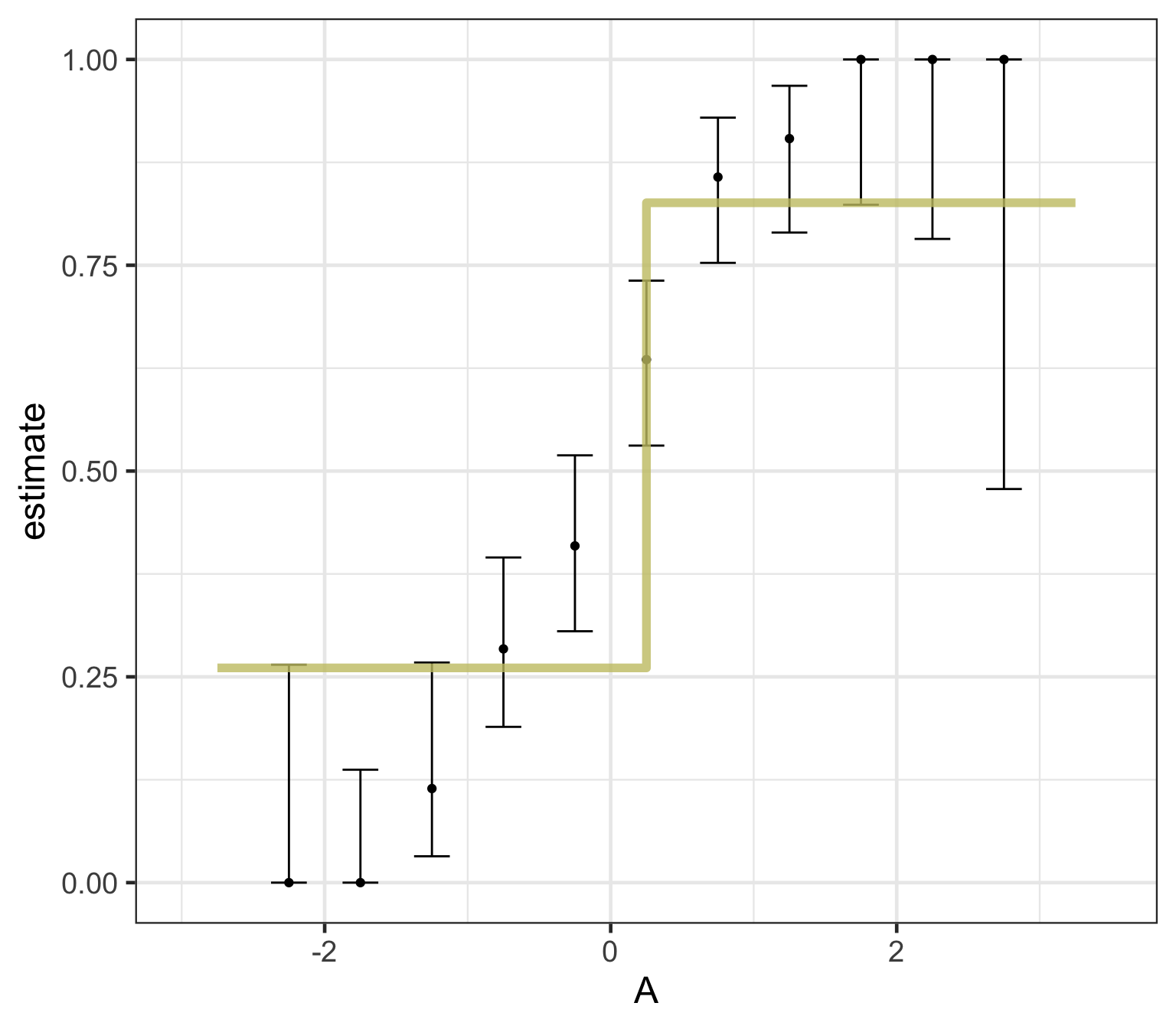
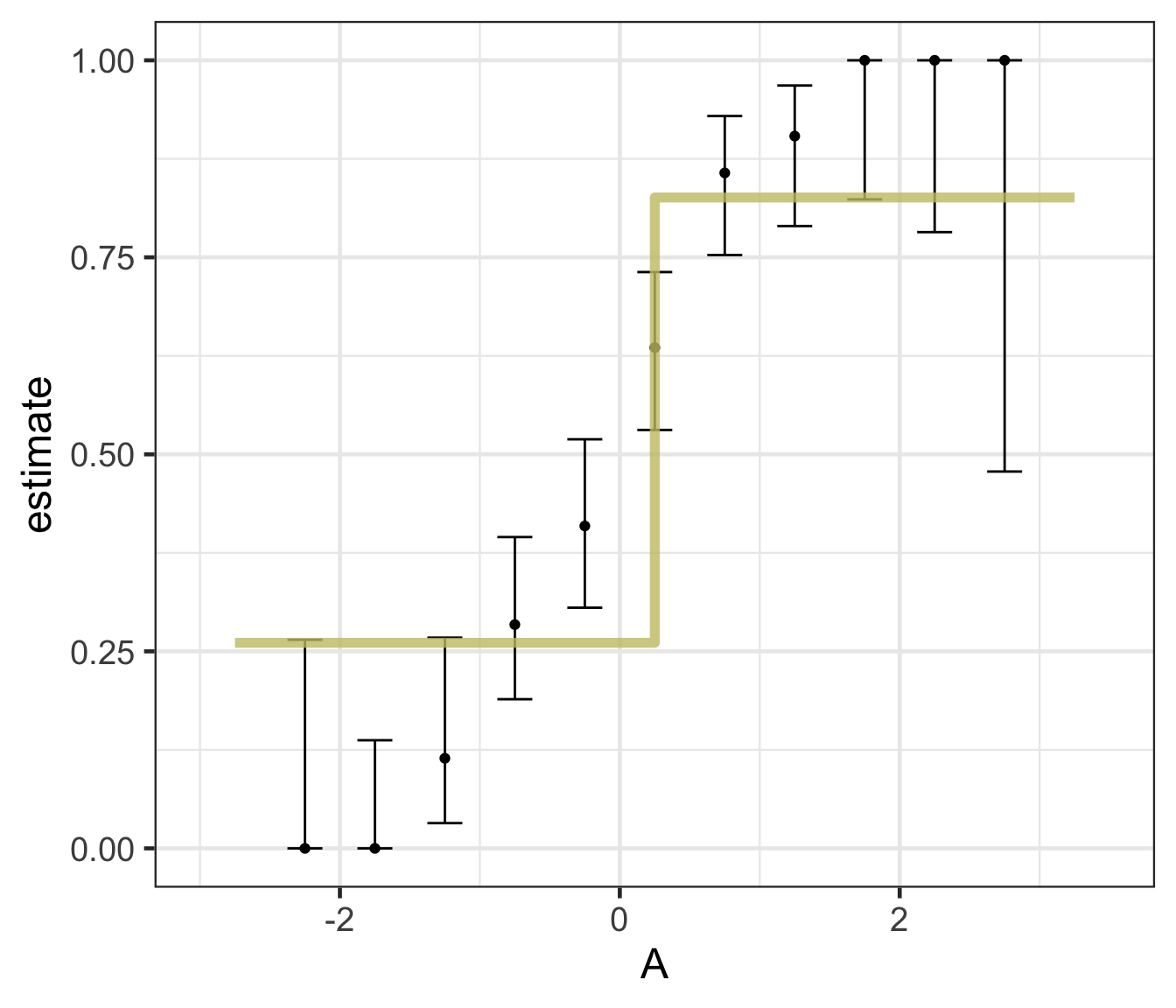
Logistic regression predicts probability—instead of a straight line, it gives an S-shaped curve that estimates how likely an outcome (e.g., is forested) is based on a predictor (e.g., rainfall and temperature).
The dots in the plot show the actual proportion of “successes” (e.g., is forested) within different bins of the predictor variable(s) (A).
The vertical error bars represent uncertainty—showing a range where the true probability might fall.
Logistic regression helps answer “how likely” questions—e.g., “How likely is someone to pass based on their study hours?” rather than just predicting a yes/no outcome.
Decision trees
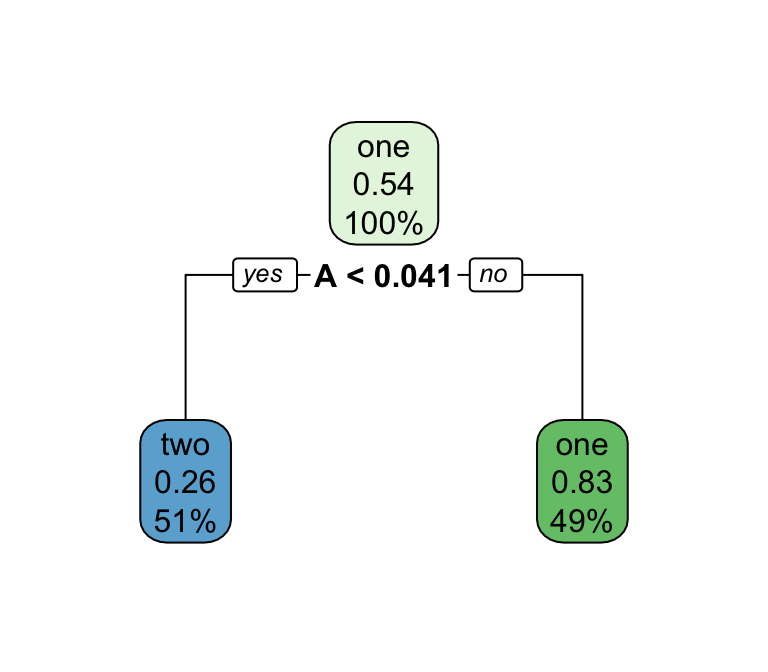
Decision trees
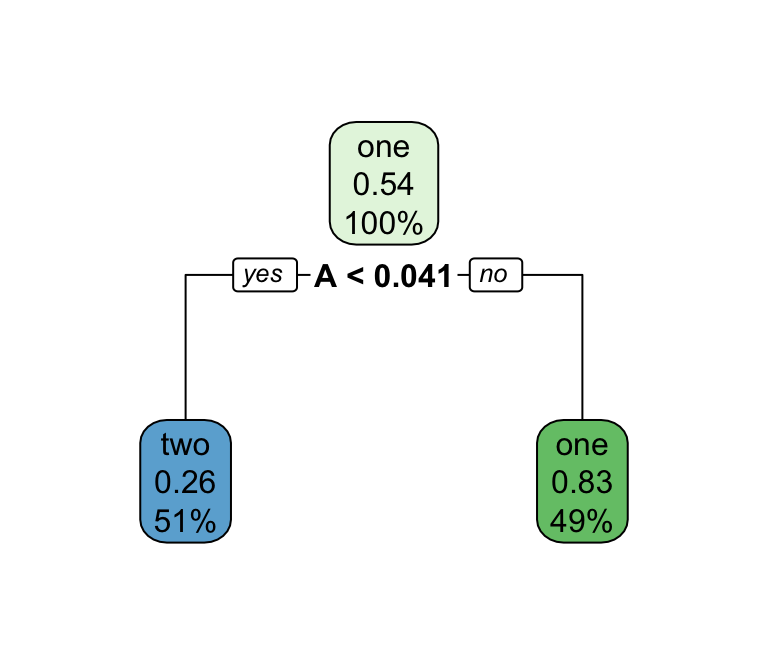
Series of splits or if/then statements based on predictors
First the tree grows until some condition is met (maximum depth, no more data)
Then the tree is pruned to reduce its complexity
Decision trees
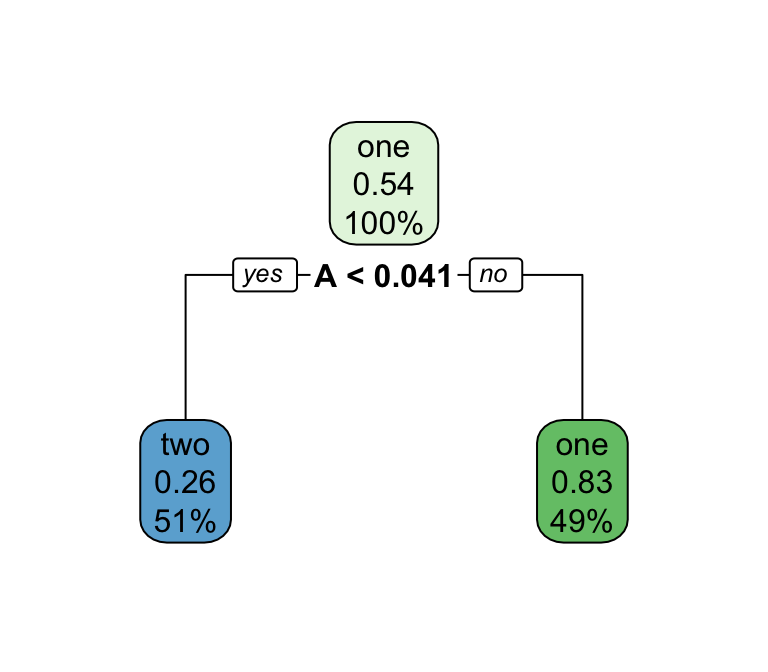
What algorithm is best for estimate forested plots?
We can only really know by testing them …
Logistic regression
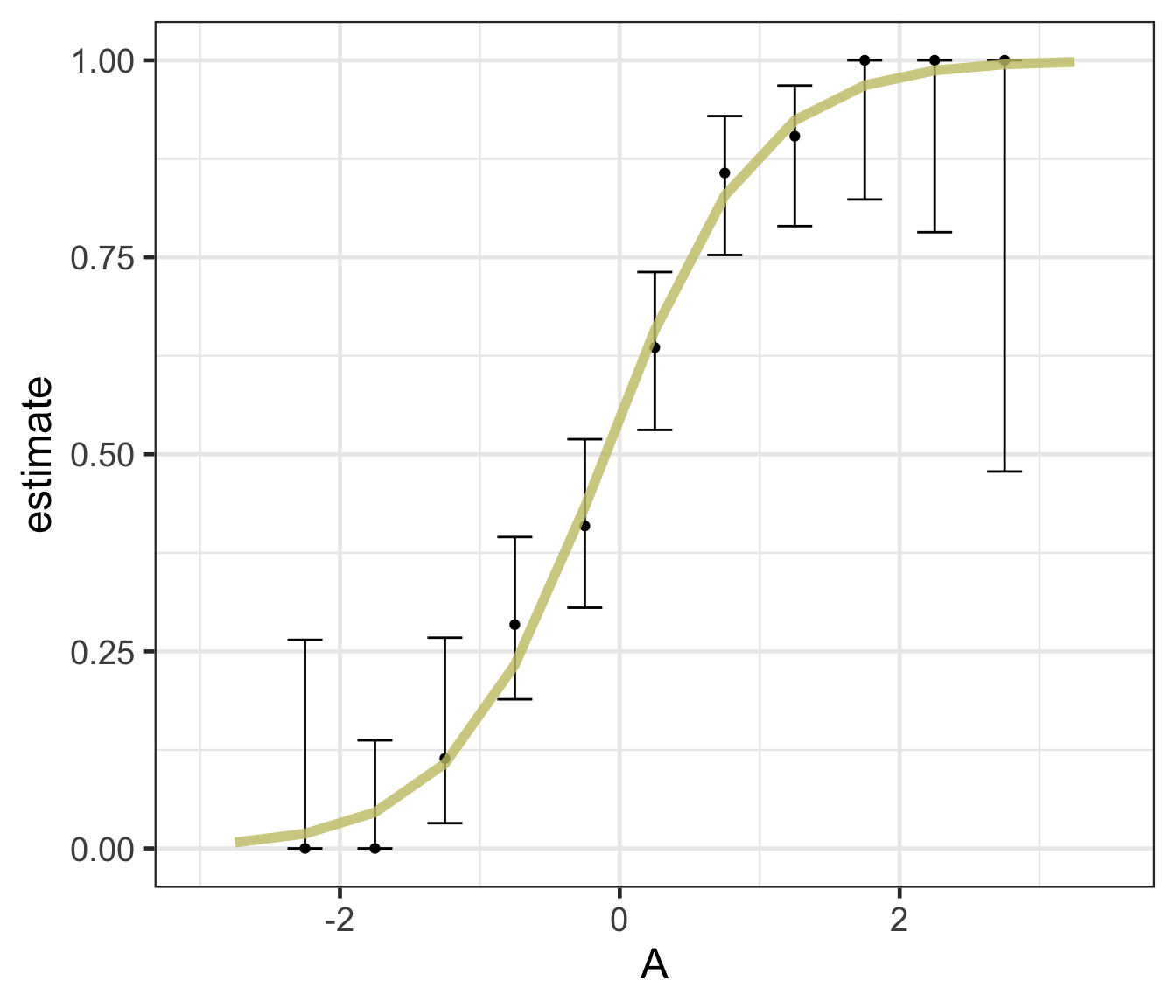
Decision trees
Fitting your model:
- OK! So you have split data, you have a model, and you have an engine and mode
- Now you need to fit the model!
- This is done using the
fit()function
dt_model <- decision_tree() %>%
set_engine('rpart') %>%
set_mode("classification")
# Model, formula, data inputs
(output <- fit(dt_model, forested ~ ., data = forested_train) )
#> parsnip model object
#>
#> n= 5685
#>
#> node), split, n, loss, yval, (yprob)
#> * denotes terminal node
#>
#> 1) root 5685 2571 Yes (0.54775726 0.45224274)
#> 2) land_type=Tree 3025 289 Yes (0.90446281 0.09553719) *
#> 3) land_type=Barren,Non-tree vegetation 2660 378 No (0.14210526 0.85789474)
#> 6) temp_annual_max< 13.395 351 155 Yes (0.55840456 0.44159544)
#> 12) tree_no_tree=Tree 91 5 Yes (0.94505495 0.05494505) *
#> 13) tree_no_tree=No tree 260 110 No (0.42307692 0.57692308) *
#> 7) temp_annual_max>=13.395 2309 182 No (0.07882200 0.92117800) *Component extracts:
While
tidymodelsprovides a standard interface to all models all base models have specific methods (remembersummary.lm? )To use these, you must extract a the underlying object.
extract_fit_engine()extracts the underlying model objectextract_fit_parsnip()extracts the parsnip objectextract_recipe()extracts the recipe objectextract_preprocessor()extracts the preprocessor object… many more
ex <- extract_fit_engine(output)
class(ex) # the class of the engine used!
#> [1] "rpart"
# Use rpart plot and text methods ...
plot(ex)
text(ex, cex = 0.9, use.n = TRUE, xpd = TRUE,)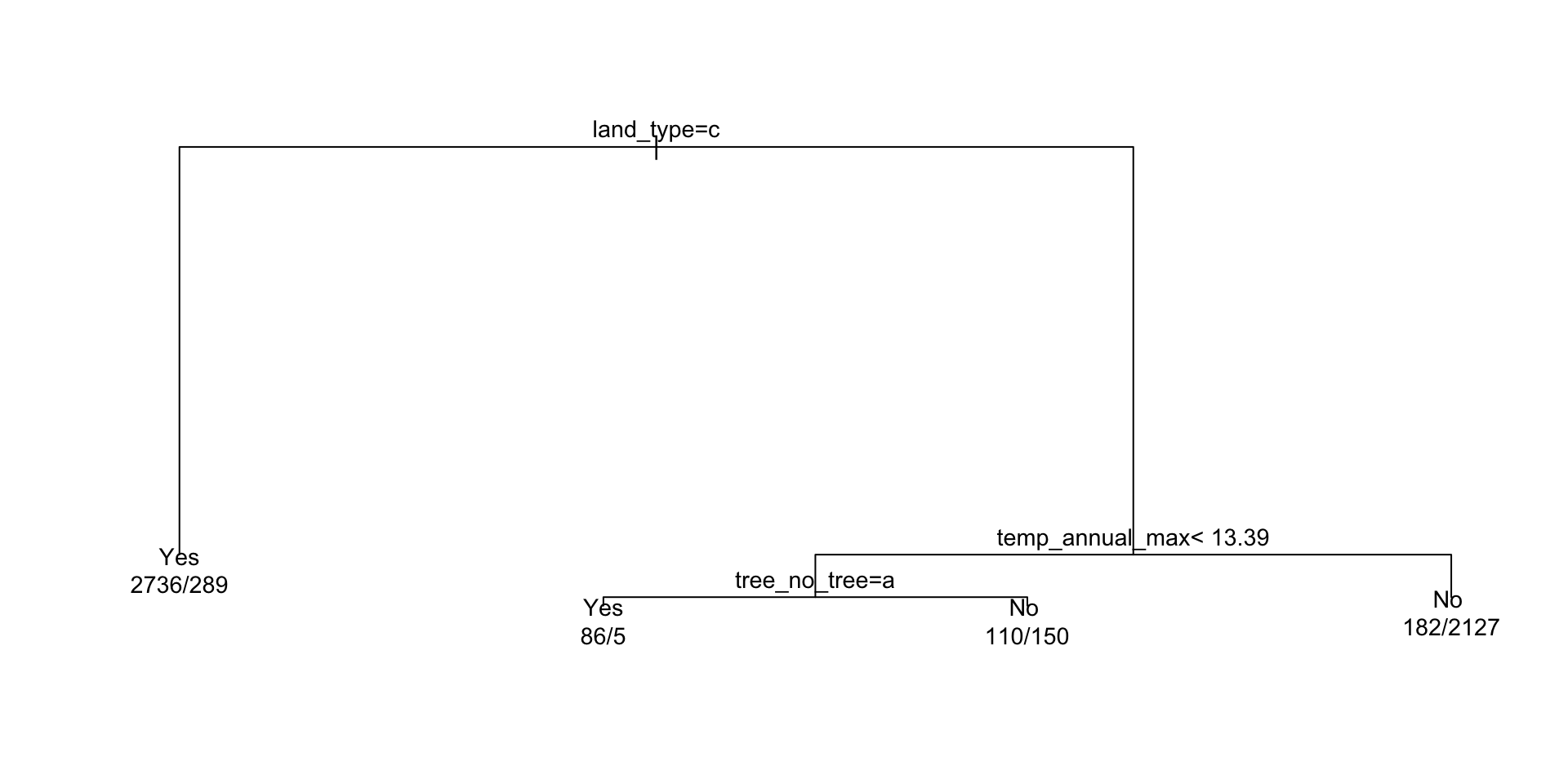
A model workflow
Workflows? 
The workflows package in tidymodels helps:
Manage preprocessing and modeling in a structured pipeline
Avoid repetitive code
Reduce errors when integrating steps
. . .
Think of them as containers for:
Preprocessing steps (e.g., feature engineering, transformations)
Model specification
Fitting approaches
. . .
Why Use Workflows?
✅ Keeps preprocessing and modeling together
✅ Ensures consistency in data handling
✅ Makes code easier to read and maintain
Basic Workflow Structure: 
Create a workflow object (similar to how
ggplot()instantiates a canvas)Add a formula or recipe (preprocessor)
Add the model
Fit the workflow
A model workflow 

dt_wf <- workflow() %>%
add_formula(forested ~ .) %>%
add_model(dt_model) %>%
fit(data = forested_train)#> ══ Workflow [trained] ══════════════════════════════════════════════════════════
#> Preprocessor: Formula
#> Model: decision_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> forested ~ .
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> n= 5685
#>
#> node), split, n, loss, yval, (yprob)
#> * denotes terminal node
#>
#> 1) root 5685 2571 Yes (0.54775726 0.45224274)
#> 2) land_type=Tree 3025 289 Yes (0.90446281 0.09553719) *
#> 3) land_type=Barren,Non-tree vegetation 2660 378 No (0.14210526 0.85789474)
#> 6) temp_annual_max< 13.395 351 155 Yes (0.55840456 0.44159544)
#> 12) tree_no_tree=Tree 91 5 Yes (0.94505495 0.05494505) *
#> 13) tree_no_tree=No tree 260 110 No (0.42307692 0.57692308) *
#> 7) temp_annual_max>=13.395 2309 182 No (0.07882200 0.92117800) *Predict with your model 
How do you use your new model?
augment()will return the dataset with predictions and residuals added.
dt_preds <- augment(dt_wf, new_data = forested_test)#> # A tibble: 1,422 × 22
#> .pred_class .pred_Yes .pred_No forested year elevation eastness northness
#> <fct> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl>
#> 1 Yes 0.904 0.0955 No 2005 164 -84 53
#> 2 Yes 0.904 0.0955 Yes 2005 806 47 -88
#> 3 No 0.423 0.577 Yes 2005 2240 -67 -74
#> 4 Yes 0.904 0.0955 Yes 2005 787 66 -74
#> 5 Yes 0.904 0.0955 Yes 2005 1330 99 7
#> 6 Yes 0.904 0.0955 Yes 2005 1423 46 88
#> 7 Yes 0.904 0.0955 Yes 2014 546 -92 -38
#> 8 Yes 0.904 0.0955 Yes 2014 1612 30 -95
#> 9 No 0.0788 0.921 No 2014 235 0 -100
#> 10 No 0.0788 0.921 No 2014 308 -70 -70
#> # ℹ 1,412 more rows
#> # ℹ 14 more variables: roughness <dbl>, tree_no_tree <fct>, dew_temp <dbl>,
#> # precip_annual <dbl>, temp_annual_mean <dbl>, temp_annual_min <dbl>,
#> # temp_annual_max <dbl>, temp_january_min <dbl>, vapor_min <dbl>,
#> # vapor_max <dbl>, canopy_cover <dbl>, lon <dbl>, lat <dbl>, land_type <fct>Evaluate your model 
How do you evaluate the skill of your models?
We will learn more about model evaluation and tuning latter in this unit, but for now we can blindly use the metrics() function defaults to get a sense of how our model is doing.
The default metrics for classification models are accuracy and kap (Cohen’s Kappa)
The default metric for regression models are RMSE, R^2, and MAE
# We have a classification model
metrics(dt_preds, truth = forested, estimate = .pred_class)
#> # A tibble: 2 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 accuracy binary 0.885
#> 2 kap binary 0.768tidymodels advantage! 
- OK! while that was not too much work, it certainly wasn’t minimal.
. . .
Now lets say you want to check if the logistic regression model performs better than the decision tree
That sounds like a lot to repeat.
Fortunately, the
tidymodelsframework makes this a straight forward swap!
. . .
log_mod = logistic_reg() %>%
set_engine("glm") %>%
set_mode("classification")
log_wf <- workflow() %>%
add_formula(forested ~ .) %>%
add_model(log_mod) %>%
fit(data = forested_train)
log_preds <- augment(log_wf, new_data = forested_test)
metrics(log_preds, truth = forested, estimate = .pred_class)
#> # A tibble: 2 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 accuracy binary 0.890
#> 2 kap binary 0.777So who wins?
metrics(dt_preds, truth = forested, estimate = .pred_class)
#> # A tibble: 2 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 accuracy binary 0.885
#> 2 kap binary 0.768
metrics(log_preds, truth = forested, estimate = .pred_class)
#> # A tibble: 2 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 accuracy binary 0.890
#> 2 kap binary 0.777Right out of the gate, there doesn’t seem to be much difference, but …
Don’t get to confident!
- We have evaluated the model on the same data we trained it on
- This is not a good practice and can give us a false sense of confidence
- Instead, we can use our
resamplesto better understand the skill of a model based on the iterative leave out policy:
dt_wf_rs <- workflow() %>%
add_formula(forested ~ .) %>%
add_model(dt_model) %>%
fit_resamples(resamples = forested_folds,
# Here we just ask tidymodels to save all predictions...
control = control_resamples(save_pred = TRUE))#> # A tibble: 1 × 5
#> splits id .metrics .notes .predictions
#> <list> <chr> <list> <list> <list>
#> 1 <split [5116/569]> Fold01 <tibble [3 × 4]> <tibble [0 × 3]> <tibble [569 × 6]>tidyr::unnest could be used to expand any of those list columns!
Don’t get to confident!
- We can execute the same workflow for the logistic regression model, this time evaluating the resamples instead of the test data.
log_wf_rs <- workflow() %>%
add_formula(forested ~ .) %>%
add_model(log_mod) %>%
fit_resamples(resamples = forested_folds,
control = control_resamples(save_pred = TRUE))#> # Resampling results
#> # 10-fold cross-validation
#> # A tibble: 10 × 5
#> splits id .metrics .notes .predictions
#> <list> <chr> <list> <list> <list>
#> 1 <split [5116/569]> Fold01 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 2 <split [5116/569]> Fold02 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 3 <split [5116/569]> Fold03 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 4 <split [5116/569]> Fold04 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 5 <split [5116/569]> Fold05 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 6 <split [5117/568]> Fold06 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 7 <split [5117/568]> Fold07 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 8 <split [5117/568]> Fold08 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 9 <split [5117/568]> Fold09 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>
#> 10 <split [5117/568]> Fold10 <tibble [3 × 4]> <tibble [0 × 3]> <tibble>Don’t get to confident!
Since we now have an “ensemble” of models (across the folds), we can collect a summary of the metrics across them.
The default metrics for classification ensembles are:
- accuracy: accuracy is the proportion of correct predictions
- brier_class: the Brier score for classification is the mean squared difference between the predicted probability and the actual outcome
- roc_auc: the area under the ROC (Receiver Operating Characteristic) curve
collect_metrics(log_wf_rs)
#> # A tibble: 3 × 6
#> .metric .estimator mean n std_err .config
#> <chr> <chr> <dbl> <int> <dbl> <chr>
#> 1 accuracy binary 0.906 10 0.00360 Preprocessor1_Model1
#> 2 brier_class binary 0.0714 10 0.00191 Preprocessor1_Model1
#> 3 roc_auc binary 0.961 10 0.00164 Preprocessor1_Model1
collect_metrics(dt_wf_rs)
#> # A tibble: 3 × 6
#> .metric .estimator mean n std_err .config
#> <chr> <chr> <dbl> <int> <dbl> <chr>
#> 1 accuracy binary 0.896 10 0.00385 Preprocessor1_Model1
#> 2 brier_class binary 0.0889 10 0.00224 Preprocessor1_Model1
#> 3 roc_auc binary 0.909 10 0.00267 Preprocessor1_Model1Further simplification: workflowsets
OK, so we have streamlined a lot of things with tidymodels:
We have a specified preprocesser (e.g. formula or recipe)
We have defined a model (complete with engine and mode)
We have implemented workflows pairing each model with the preprocesser to either fit the model to the resamples, or, the training data.
While a significant improvement, we can do better!
Does the idea of implementing a process over an list of elements ring a bell?
Model mappings: 
workflowsetsis a package that builds off ofpurrrand allows you to iterate over multiple models and/or multiple resamplesRemember how
map,map_*,map2, andwalk2functions allow lists to map to lists or vectors?workflow_setmaps a preprocessor (formula or recipe) to a set of models - each provided as alistobject.To start, we will create a
workflow_setobject (instead of aworkflow). The first argument is a list of preprocessor objects (formulas or recipes) and the second argument is a list of model objects.Both must be lists, by nature of the underlying
purrrcode.
(wf_obj <- workflow_set(list(forested ~.), list(log_mod, dt_model)))
#> # A workflow set/tibble: 2 × 4
#> wflow_id info option result
#> <chr> <list> <list> <list>
#> 1 formula_logistic_reg <tibble [1 × 4]> <opts[0]> <list [0]>
#> 2 formula_decision_tree <tibble [1 × 4]> <opts[0]> <list [0]>Iteritive extecution
Once the
workflow_setobject is created, theworkflow_mapfunction can be used to map a function across the preprocessor/model combinations.Here we are mapping the
fit_resamplesfunction across theworkflow_setcombinations using theresamplesargument to specify the resamples we want to use (forested_folds).
wf_obj <-
workflow_set(list(forested ~.), list(log_mod, dt_model)) %>%
workflow_map("fit_resamples", resamples = forested_folds) #> # A workflow set/tibble: 2 × 4
#> wflow_id info option result
#> <chr> <list> <list> <list>
#> 1 formula_logistic_reg <tibble [1 × 4]> <opts[1]> <rsmp[+]>
#> 2 formula_decision_tree <tibble [1 × 4]> <opts[1]> <rsmp[+]>Rapid comparision
With that single function, all models have been fit to the resamples and we can quickly compare the results both graphically and statistically:
# Quick plot function
autoplot(wf_obj) +
theme_linedraw()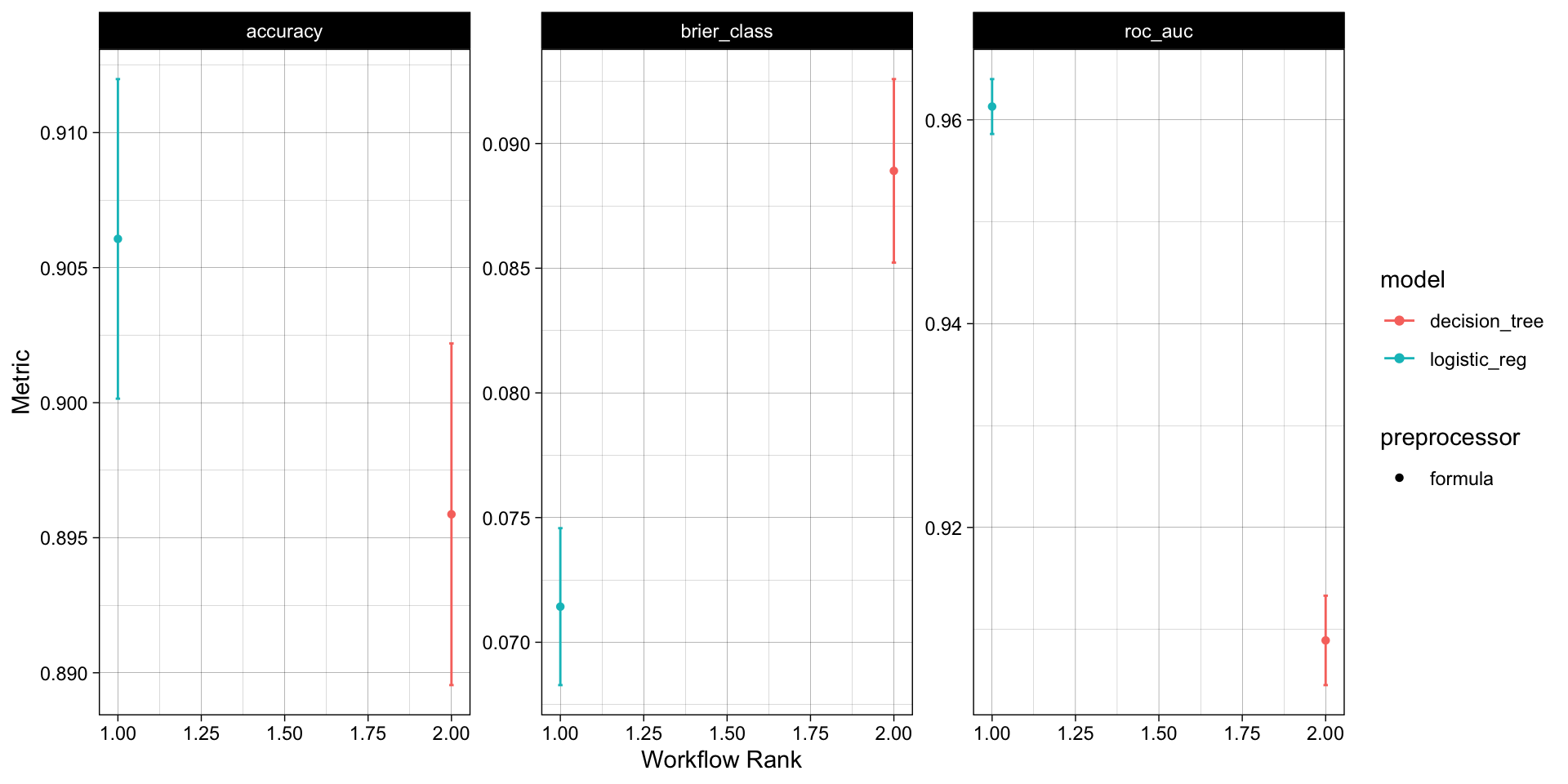
# Long list of results, ranked by accuracy
rank_results(wf_obj,
rank_metric = "accuracy",
select_best = TRUE)
#> # A tibble: 6 × 9
#> wflow_id .config .metric mean std_err n preprocessor model rank
#> <chr> <chr> <chr> <dbl> <dbl> <int> <chr> <chr> <int>
#> 1 formula_logisti… Prepro… accura… 0.906 0.00360 10 formula logi… 1
#> 2 formula_logisti… Prepro… brier_… 0.0714 0.00191 10 formula logi… 1
#> 3 formula_logisti… Prepro… roc_auc 0.961 0.00164 10 formula logi… 1
#> 4 formula_decisio… Prepro… accura… 0.896 0.00385 10 formula deci… 2
#> 5 formula_decisio… Prepro… brier_… 0.0889 0.00224 10 formula deci… 2
#> 6 formula_decisio… Prepro… roc_auc 0.909 0.00267 10 formula deci… 2. . .
Overall, the logistic regression model appears to the best model for this data set!
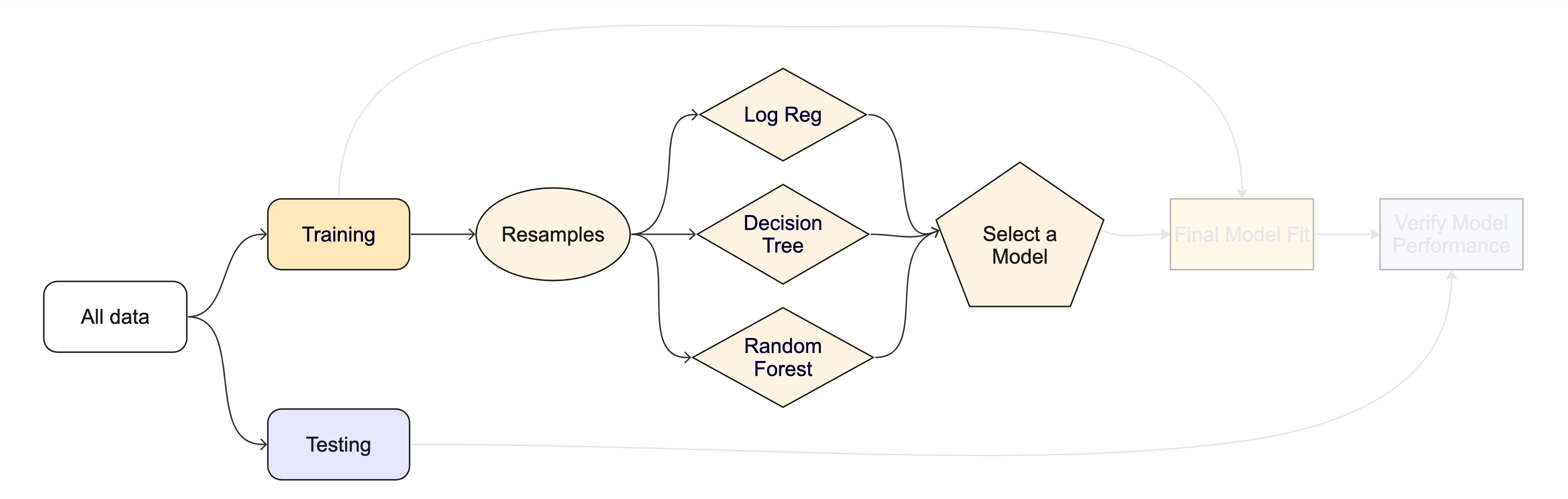
The whole game - status update
On Wednesday, we’ll talk more about models we can chose from…
Next week we will cover